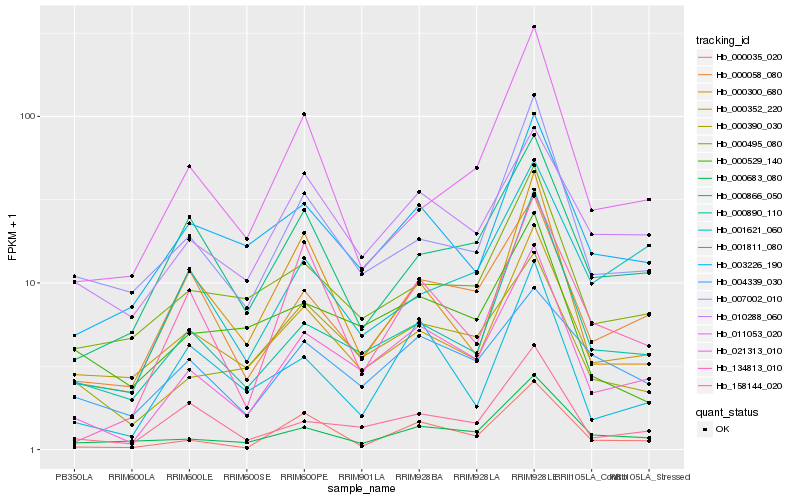
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_021313_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0010s24380g [Populus trichocarpa] |
| 2 |
Hb_000890_110 |
0.1434168517 |
- |
- |
PREDICTED: GTP-binding protein OBGC, chloroplastic [Populus euphratica] |
| 3 |
Hb_158144_020 |
0.1441710473 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Vitis vinifera] |
| 4 |
Hb_000035_020 |
0.1512057774 |
- |
- |
PREDICTED: omega-hydroxypalmitate O-feruloyl transferase [Jatropha curcas] |
| 5 |
Hb_000058_080 |
0.1525822206 |
- |
- |
PREDICTED: phenylalanine--tRNA ligase, chloroplastic/mitochondrial [Jatropha curcas] |
| 6 |
Hb_000683_080 |
0.1536282467 |
- |
- |
hypothetical protein EUGRSUZ_G013602, partial [Eucalyptus grandis] |
| 7 |
Hb_000390_030 |
0.1544415209 |
- |
- |
hypothetical protein ZEAMMB73_364791 [Zea mays] |
| 8 |
Hb_007002_010 |
0.1554704396 |
- |
- |
PREDICTED: dnaJ protein ERDJ3B [Jatropha curcas] |
| 9 |
Hb_003226_190 |
0.1607365439 |
- |
- |
PREDICTED: blue-light photoreceptor PHR2 [Jatropha curcas] |
| 10 |
Hb_000352_220 |
0.1653603185 |
- |
- |
PREDICTED: probable tyrosine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 11 |
Hb_000495_080 |
0.1655418075 |
- |
- |
Pentatricopeptide repeat (PPR-like) superfamily protein [Theobroma cacao] |
| 12 |
Hb_010288_060 |
0.1687027173 |
- |
- |
PREDICTED: dnaJ protein ERDJ3B [Jatropha curcas] |
| 13 |
Hb_001811_080 |
0.1771484703 |
- |
- |
PREDICTED: fructose-1,6-bisphosphatase, cytosolic [Jatropha curcas] |
| 14 |
Hb_134813_010 |
0.1775690906 |
- |
- |
aldo/keto reductase, putative [Ricinus communis] |
| 15 |
Hb_000866_050 |
0.1781225716 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 16 |
Hb_000529_140 |
0.1833263245 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000300_680 |
0.1856579242 |
- |
- |
PREDICTED: uncharacterized protein LOC105632625 isoform X1 [Jatropha curcas] |
| 18 |
Hb_011053_020 |
0.1857831785 |
- |
- |
lipoic acid synthetase, putative [Ricinus communis] |
| 19 |
Hb_001621_060 |
0.1865465812 |
- |
- |
PREDICTED: thylakoid lumenal 15 kDa protein 1, chloroplastic [Jatropha curcas] |
| 20 |
Hb_004339_030 |
0.189108233 |
- |
- |
PREDICTED: 7-methylguanosine phosphate-specific 5'-nucleotidase A isoform X2 [Jatropha curcas] |