| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
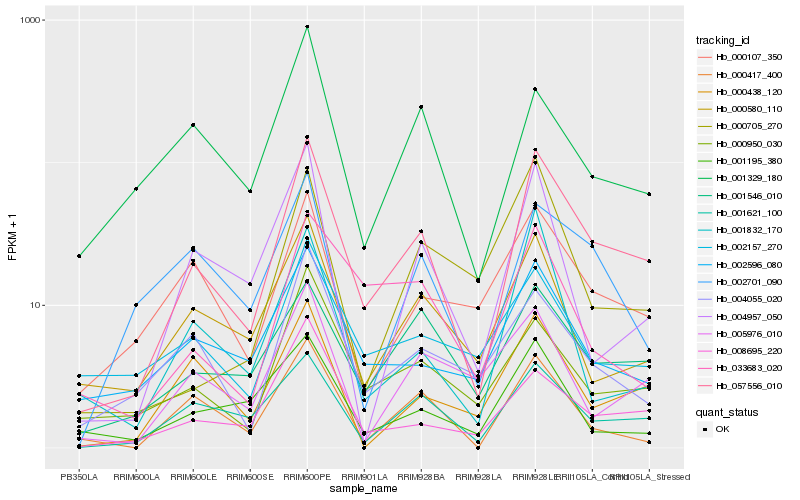
Hb_057556_010 |
0.0 |
- |
- |
Light-regulated protein precursor, putative [Ricinus communis] |
| 2 |
Hb_001621_100 |
0.1673210825 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase S.4 [Jatropha curcas] |
| 3 |
Hb_001546_010 |
0.1905711177 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_008695_220 |
0.1929262913 |
- |
- |
PREDICTED: uncharacterized protein LOC105638863 [Jatropha curcas] |
| 5 |
Hb_000950_030 |
0.1945615709 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g10290 [Nelumbo nucifera] |
| 6 |
Hb_000580_110 |
0.1979482216 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 3 [Jatropha curcas] |
| 7 |
Hb_000417_400 |
0.1993949651 |
- |
- |
PREDICTED: ubiquitin domain-containing protein 1-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_002596_080 |
0.2000595736 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At5g04160 [Jatropha curcas] |
| 9 |
Hb_000438_120 |
0.2011650318 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 10 |
Hb_000107_350 |
0.2023258917 |
- |
- |
PREDICTED: uncharacterized protein LOC105635576 [Jatropha curcas] |
| 11 |
Hb_004957_050 |
0.2046415194 |
- |
- |
hypothetical protein POPTR_0010s12360g [Populus trichocarpa] |
| 12 |
Hb_004055_020 |
0.2109543361 |
- |
- |
PREDICTED: uncharacterized protein LOC105631433 [Jatropha curcas] |
| 13 |
Hb_033683_020 |
0.212069425 |
- |
- |
hypothetical protein POPTR_1072s00200g [Populus trichocarpa] |
| 14 |
Hb_001195_380 |
0.2163154752 |
- |
- |
PREDICTED: uncharacterized protein LOC105633781 [Jatropha curcas] |
| 15 |
Hb_002701_090 |
0.2165293024 |
- |
- |
hypothetical protein POPTR_0017s13430g [Populus trichocarpa] |
| 16 |
Hb_005976_010 |
0.2169816361 |
- |
- |
PREDICTED: PI-PLC X domain-containing protein At5g67130-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_001832_170 |
0.2182706469 |
- |
- |
PREDICTED: beta-galactosidase-like [Jatropha curcas] |
| 18 |
Hb_002157_270 |
0.21871729 |
- |
- |
PREDICTED: transmembrane protein 87B [Jatropha curcas] |
| 19 |
Hb_001329_180 |
0.2202347457 |
- |
- |
S-adenosylmethionine synthetase 1 family protein [Populus trichocarpa] |
| 20 |
Hb_000705_270 |
0.2213833335 |
- |
- |
PREDICTED: uncharacterized protein LOC105131117 [Populus euphratica] |