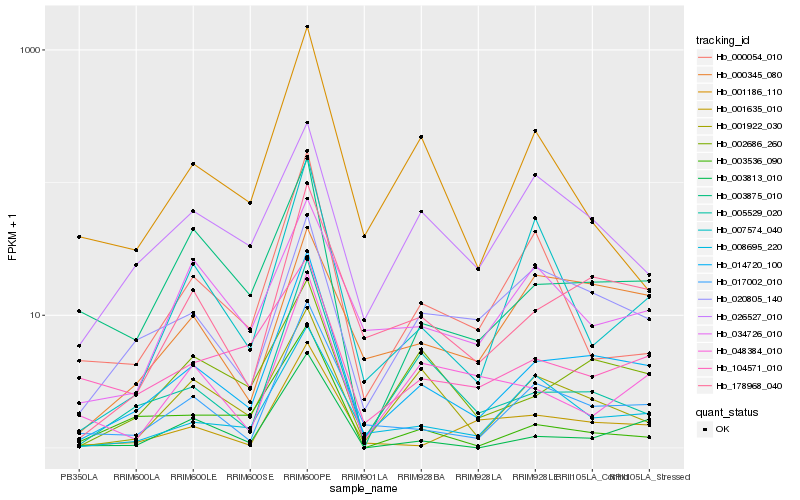
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_014720_100 |
0.0 |
- |
- |
PREDICTED: probable boron transporter 2 [Jatropha curcas] |
| 2 |
Hb_178968_040 |
0.1267000703 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_017002_010 |
0.1423126633 |
- |
- |
PREDICTED: expansin-A15-like [Jatropha curcas] |
| 4 |
Hb_008695_220 |
0.1796857317 |
- |
- |
PREDICTED: uncharacterized protein LOC105638863 [Jatropha curcas] |
| 5 |
Hb_005529_020 |
0.1809520928 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 32 [Jatropha curcas] |
| 6 |
Hb_002686_260 |
0.1832255998 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein 21/22, putative [Ricinus communis] |
| 7 |
Hb_003875_010 |
0.1883680015 |
- |
- |
hypothetical protein POPTR_0002s14490g, partial [Populus trichocarpa] |
| 8 |
Hb_001635_010 |
0.1889661433 |
- |
- |
- |
| 9 |
Hb_003813_010 |
0.19071267 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor BEE 1-like [Jatropha curcas] |
| 10 |
Hb_001922_030 |
0.2078092843 |
- |
- |
PREDICTED: probable membrane-associated kinase regulator 5 [Jatropha curcas] |
| 11 |
Hb_020805_140 |
0.212400465 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At5g42360-like [Jatropha curcas] |
| 12 |
Hb_007574_040 |
0.2133375134 |
- |
- |
PREDICTED: uncharacterized protein LOC105638144 [Jatropha curcas] |
| 13 |
Hb_104571_010 |
0.2170901858 |
- |
- |
PREDICTED: metal transporter Nramp1 [Jatropha curcas] |
| 14 |
Hb_000054_010 |
0.2175981832 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 15 |
Hb_026527_010 |
0.2239071891 |
- |
- |
PREDICTED: caffeoylshikimate esterase-like [Populus euphratica] |
| 16 |
Hb_003536_090 |
0.2244939568 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 17 |
Hb_048384_010 |
0.2254855745 |
- |
- |
PREDICTED: probable receptor-like protein kinase At2g42960 [Cucumis sativus] |
| 18 |
Hb_034726_010 |
0.2255556188 |
- |
- |
PREDICTED: chaperone protein dnaJ 1, mitochondrial isoform X2 [Jatropha curcas] |
| 19 |
Hb_001186_110 |
0.2290599745 |
- |
- |
hypothetical protein JCGZ_18389 [Jatropha curcas] |
| 20 |
Hb_000345_080 |
0.2302690359 |
- |
- |
PREDICTED: protein RALF-like 33 [Jatropha curcas] |