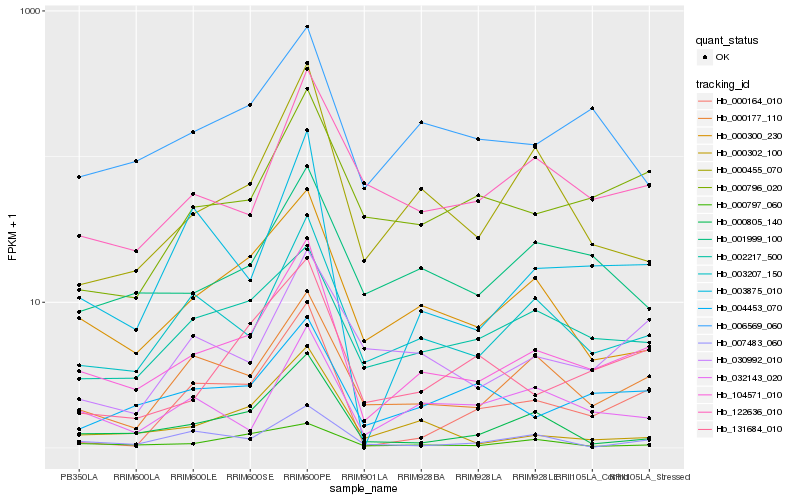
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_104571_010 |
0.0 |
- |
- |
PREDICTED: metal transporter Nramp1 [Jatropha curcas] |
| 2 |
Hb_003207_150 |
0.1471174511 |
- |
- |
PREDICTED: SPX domain-containing membrane protein At4g22990-like [Jatropha curcas] |
| 3 |
Hb_000300_230 |
0.1582690233 |
- |
- |
PREDICTED: cation/calcium exchanger 2 [Jatropha curcas] |
| 4 |
Hb_000177_110 |
0.1610737848 |
- |
- |
PREDICTED: uncharacterized protein LOC105642063 [Jatropha curcas] |
| 5 |
Hb_003875_010 |
0.1759480903 |
- |
- |
hypothetical protein POPTR_0002s14490g, partial [Populus trichocarpa] |
| 6 |
Hb_002217_500 |
0.1794659868 |
- |
- |
hypothetical protein JCGZ_19014 [Jatropha curcas] |
| 7 |
Hb_000805_140 |
0.1798136211 |
transcription factor |
TF Family: GRAS |
Chitin-inducible gibberellin-responsive protein, putative [Ricinus communis] |
| 8 |
Hb_131684_010 |
0.1798438869 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000455_070 |
0.1816307466 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: carboxymethylenebutenolidase homolog [Jatropha curcas] |
| 10 |
Hb_122636_010 |
0.1820854819 |
- |
- |
dienelactone hydrolase family protein [Populus trichocarpa] |
| 11 |
Hb_000796_020 |
0.1837553621 |
- |
- |
ADP-ribosylation factor [Phaseolus vulgaris] |
| 12 |
Hb_004453_070 |
0.18428694 |
transcription factor |
TF Family: CPP |
PREDICTED: CRC domain-containing protein TSO1-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_000302_100 |
0.1845885954 |
- |
- |
PREDICTED: uncharacterized protein LOC105766209 [Gossypium raimondii] |
| 14 |
Hb_006569_060 |
0.1864258659 |
- |
- |
plasma membrane intrinsic protein PIP2;3 [Hevea brasiliensis] |
| 15 |
Hb_001999_100 |
0.1878321737 |
- |
- |
PREDICTED: UDP-glycosyltransferase 71D1-like [Jatropha curcas] |
| 16 |
Hb_000797_060 |
0.1918118487 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 17 |
Hb_032143_020 |
0.1921347486 |
- |
- |
hypothetical protein POPTR_0009s06310g [Populus trichocarpa] |
| 18 |
Hb_007483_060 |
0.1971612009 |
desease resistance |
Gene Name: ABC_membrane |
multidrug resistance protein 1, 2, putative [Ricinus communis] |
| 19 |
Hb_030992_010 |
0.19735174 |
- |
- |
acetylornithine aminotransferase, putative [Ricinus communis] |
| 20 |
Hb_000164_010 |
0.1987044117 |
- |
- |
small heat-shock protein, putative [Ricinus communis] |