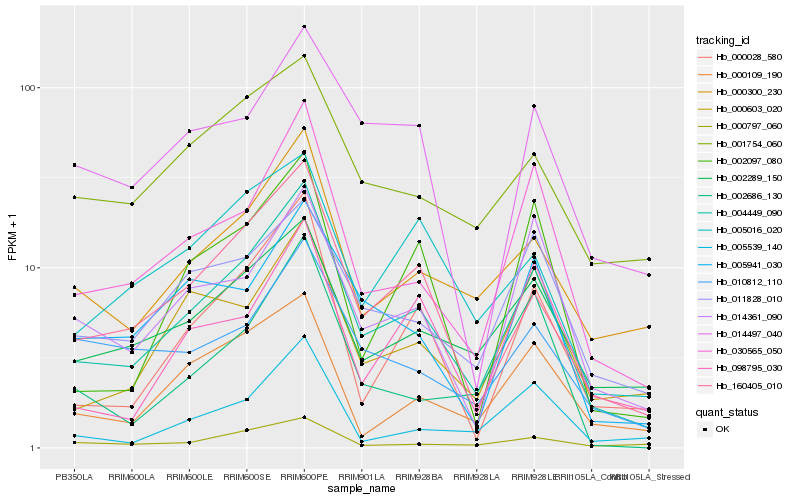
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004449_090 |
0.0 |
- |
- |
PREDICTED: CO(2)-response secreted protease-like [Jatropha curcas] |
| 2 |
Hb_160405_010 |
0.1107960908 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 3 |
Hb_030565_050 |
0.1159595713 |
- |
- |
UDP-D-GLUCURONATE 4-EPIMERASE 6 family protein [Populus trichocarpa] |
| 4 |
Hb_000028_580 |
0.1345101121 |
- |
- |
PREDICTED: glutamate decarboxylase 5-like [Jatropha curcas] |
| 5 |
Hb_000300_230 |
0.1348098129 |
- |
- |
PREDICTED: cation/calcium exchanger 2 [Jatropha curcas] |
| 6 |
Hb_002289_150 |
0.1414226339 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 3 [UDP-forming]-like [Jatropha curcas] |
| 7 |
Hb_011828_010 |
0.1544718979 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g67720 [Jatropha curcas] |
| 8 |
Hb_000109_190 |
0.1559589998 |
- |
- |
PREDICTED: RING-H2 finger protein ATL47 [Jatropha curcas] |
| 9 |
Hb_000603_020 |
0.1562904578 |
- |
- |
PREDICTED: uncharacterized protein LOC105638063 isoform X1 [Jatropha curcas] |
| 10 |
Hb_014361_090 |
0.1571244629 |
- |
- |
acyl-CoA binding protein 3A [Vernicia fordii] |
| 11 |
Hb_005941_030 |
0.1584591295 |
transcription factor |
TF Family: HB |
DNA binding protein, putative [Ricinus communis] |
| 12 |
Hb_005016_020 |
0.1597335603 |
desease resistance |
Gene Name: ABC_membrane |
ABC transporter family protein [Hevea brasiliensis] |
| 13 |
Hb_002097_080 |
0.161021677 |
- |
- |
phosphatidylinositol-4-phosphate 5-kinase, putative [Ricinus communis] |
| 14 |
Hb_005539_140 |
0.1641850275 |
- |
- |
PREDICTED: U-box domain-containing protein 19-like [Jatropha curcas] |
| 15 |
Hb_014497_040 |
0.164232746 |
- |
- |
PREDICTED: calvin cycle protein CP12-3, chloroplastic [Jatropha curcas] |
| 16 |
Hb_001754_060 |
0.1651028558 |
- |
- |
Xyloglucan 6-xylosyltransferase, putative [Ricinus communis] |
| 17 |
Hb_002686_130 |
0.1672405793 |
transcription factor |
TF Family: G2-like |
PREDICTED: uncharacterized protein LOC105650401 [Jatropha curcas] |
| 18 |
Hb_010812_110 |
0.1689196818 |
- |
- |
putative plant disease resistance family protein [Populus trichocarpa] |
| 19 |
Hb_000797_060 |
0.1690777909 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 20 |
Hb_098795_030 |
0.1698587143 |
- |
- |
PREDICTED: metal tolerance protein 1-like [Vitis vinifera] |