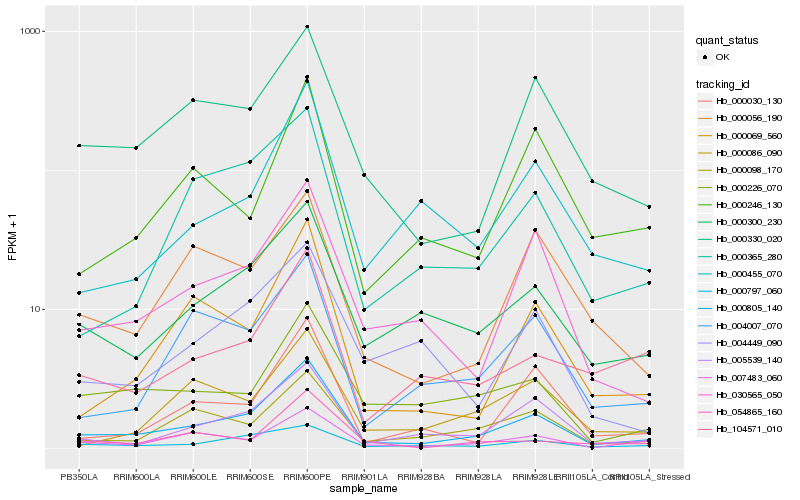
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000805_140 |
0.0 |
transcription factor |
TF Family: GRAS |
Chitin-inducible gibberellin-responsive protein, putative [Ricinus communis] |
| 2 |
Hb_005539_140 |
0.1473892667 |
- |
- |
PREDICTED: U-box domain-containing protein 19-like [Jatropha curcas] |
| 3 |
Hb_000300_230 |
0.1528511564 |
- |
- |
PREDICTED: cation/calcium exchanger 2 [Jatropha curcas] |
| 4 |
Hb_030565_050 |
0.1608485892 |
- |
- |
UDP-D-GLUCURONATE 4-EPIMERASE 6 family protein [Populus trichocarpa] |
| 5 |
Hb_000797_060 |
0.1616188657 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 6 |
Hb_007483_060 |
0.1629167149 |
desease resistance |
Gene Name: ABC_membrane |
multidrug resistance protein 1, 2, putative [Ricinus communis] |
| 7 |
Hb_000330_020 |
0.164303347 |
- |
- |
PREDICTED: isoflavone reductase-like protein [Jatropha curcas] |
| 8 |
Hb_000098_170 |
0.1710880347 |
- |
- |
hypothetical protein JCGZ_06491 [Jatropha curcas] |
| 9 |
Hb_000069_560 |
0.1713669365 |
- |
- |
hypothetical protein POPTR_0002s16980g [Populus trichocarpa] |
| 10 |
Hb_000365_280 |
0.1719256462 |
- |
- |
calcium binding protein/cast, putative [Ricinus communis] |
| 11 |
Hb_000030_130 |
0.1772206206 |
transcription factor |
TF Family: G2-like |
transcription factor, putative [Ricinus communis] |
| 12 |
Hb_004007_070 |
0.1784833792 |
- |
- |
adenylsulfate kinase, putative [Ricinus communis] |
| 13 |
Hb_000226_070 |
0.1797604195 |
- |
- |
PREDICTED: pyrophosphate-energized vacuolar membrane proton pump [Jatropha curcas] |
| 14 |
Hb_104571_010 |
0.1798136211 |
- |
- |
PREDICTED: metal transporter Nramp1 [Jatropha curcas] |
| 15 |
Hb_000455_070 |
0.1800891518 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: carboxymethylenebutenolidase homolog [Jatropha curcas] |
| 16 |
Hb_000246_130 |
0.1930326642 |
- |
- |
tubulin alpha chain, putative [Ricinus communis] |
| 17 |
Hb_054865_160 |
0.1936464723 |
- |
- |
PREDICTED: D-arabinono-1,4-lactone oxidase-like [Jatropha curcas] |
| 18 |
Hb_004449_090 |
0.1940419884 |
- |
- |
PREDICTED: CO(2)-response secreted protease-like [Jatropha curcas] |
| 19 |
Hb_000056_190 |
0.1962675054 |
- |
- |
hypothetical protein [Ricinus communis] |
| 20 |
Hb_000086_090 |
0.1964268476 |
- |
- |
PREDICTED: U-box domain-containing protein 3 [Jatropha curcas] |