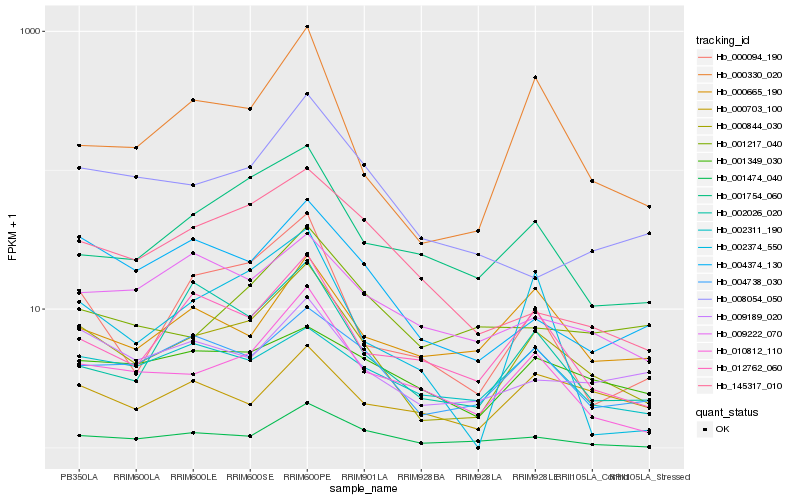
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000844_030 |
0.0 |
- |
- |
ent-kaurene oxidase, putative [Ricinus communis] |
| 2 |
Hb_010812_110 |
0.1409471434 |
- |
- |
putative plant disease resistance family protein [Populus trichocarpa] |
| 3 |
Hb_000330_020 |
0.1551861166 |
- |
- |
PREDICTED: isoflavone reductase-like protein [Jatropha curcas] |
| 4 |
Hb_004738_030 |
0.1606018024 |
- |
- |
Inactive protein kinase [Gossypium arboreum] |
| 5 |
Hb_001474_040 |
0.1662965918 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 [Jatropha curcas] |
| 6 |
Hb_012762_060 |
0.1702560323 |
- |
- |
Vitellogenic carboxypeptidase, putative [Ricinus communis] |
| 7 |
Hb_000094_190 |
0.1744051965 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 7-like [Jatropha curcas] |
| 8 |
Hb_004374_130 |
0.1802147498 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHA1B-like [Jatropha curcas] |
| 9 |
Hb_001754_060 |
0.1817710094 |
- |
- |
Xyloglucan 6-xylosyltransferase, putative [Ricinus communis] |
| 10 |
Hb_009189_020 |
0.1826386192 |
- |
- |
PREDICTED: CASP-like protein 4B4 [Jatropha curcas] |
| 11 |
Hb_000665_190 |
0.1962665289 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002374_550 |
0.1980525239 |
- |
- |
PREDICTED: aspartic proteinase nepenthesin-1 [Jatropha curcas] |
| 13 |
Hb_002311_190 |
0.1996762985 |
- |
- |
PREDICTED: uncharacterized protein LOC105643493 [Jatropha curcas] |
| 14 |
Hb_001349_030 |
0.200243951 |
- |
- |
PREDICTED: uncharacterized protein LOC105641975 isoform X1 [Jatropha curcas] |
| 15 |
Hb_002026_020 |
0.2007766756 |
- |
- |
hypothetical protein POPTR_0012s129501g, partial [Populus trichocarpa] |
| 16 |
Hb_145317_010 |
0.201991285 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 11 [Jatropha curcas] |
| 17 |
Hb_001217_040 |
0.2022295087 |
- |
- |
hypothetical protein MIMGU_mgv1a0002421mg, partial [Erythranthe guttata] |
| 18 |
Hb_000703_100 |
0.2031598765 |
- |
- |
PREDICTED: bifunctional riboflavin kinase/FMN phosphatase-like [Jatropha curcas] |
| 19 |
Hb_009222_070 |
0.203451143 |
- |
- |
PREDICTED: xylosyltransferase 2 [Jatropha curcas] |
| 20 |
Hb_008054_050 |
0.2035206248 |
- |
- |
PREDICTED: uncharacterized protein LOC101212042 [Cucumis sativus] |