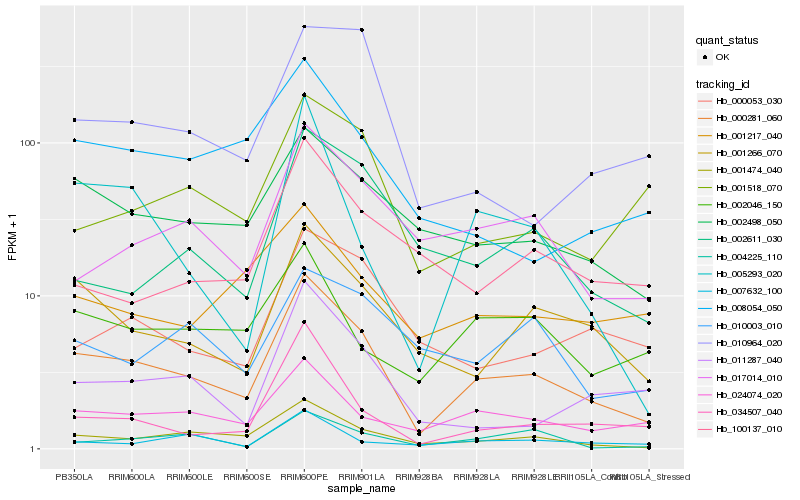
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000281_060 |
0.0 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 2 |
Hb_001474_040 |
0.1456454334 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 [Jatropha curcas] |
| 3 |
Hb_001266_070 |
0.1665884509 |
- |
- |
PREDICTED: mannan endo-1,4-beta-mannosidase 7 [Jatropha curcas] |
| 4 |
Hb_002498_050 |
0.1738215316 |
- |
- |
PREDICTED: aspartic proteinase-like [Jatropha curcas] |
| 5 |
Hb_024074_020 |
0.185507701 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g09600 [Jatropha curcas] |
| 6 |
Hb_017014_010 |
0.193946648 |
- |
- |
PREDICTED: syntaxin-132-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_034507_040 |
0.200002479 |
- |
- |
peptide transporter, putative [Ricinus communis] |
| 8 |
Hb_011287_040 |
0.2025008251 |
- |
- |
PREDICTED: OTU domain-containing protein At3g57810-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_008054_050 |
0.2062525359 |
- |
- |
PREDICTED: uncharacterized protein LOC101212042 [Cucumis sativus] |
| 10 |
Hb_001518_070 |
0.206399464 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000053_030 |
0.2068124681 |
- |
- |
hypothetical protein CISIN_1g0010393mg, partial [Citrus sinensis] |
| 12 |
Hb_004225_110 |
0.2071432914 |
- |
- |
PREDICTED: ABC transporter G family member 28 [Jatropha curcas] |
| 13 |
Hb_002046_150 |
0.2080878006 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g15080 [Jatropha curcas] |
| 14 |
Hb_001217_040 |
0.2119344313 |
- |
- |
hypothetical protein MIMGU_mgv1a0002421mg, partial [Erythranthe guttata] |
| 15 |
Hb_005293_020 |
0.212218834 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002611_030 |
0.2138107764 |
- |
- |
PREDICTED: alpha-1,3-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase [Jatropha curcas] |
| 17 |
Hb_010964_020 |
0.2166037307 |
- |
- |
RNA helicase [Arabidopsis thaliana] |
| 18 |
Hb_010003_010 |
0.2176775318 |
- |
- |
PREDICTED: uncharacterized protein LOC105630174 isoform X2 [Jatropha curcas] |
| 19 |
Hb_007632_100 |
0.2178302962 |
transcription factor |
TF Family: NF-YB |
ccaat-binding transcription factor subunit A, putative [Ricinus communis] |
| 20 |
Hb_100137_010 |
0.2202427746 |
- |
- |
hypothetical protein JCGZ_17848 [Jatropha curcas] |