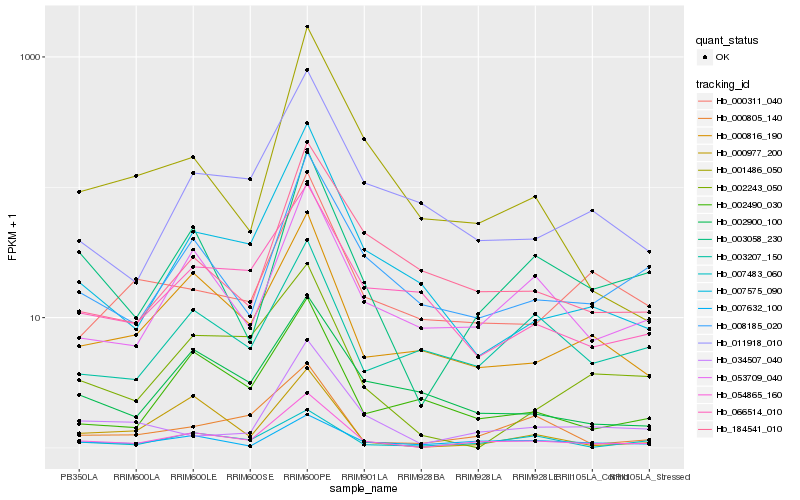
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_054865_160 |
0.0 |
- |
- |
PREDICTED: D-arabinono-1,4-lactone oxidase-like [Jatropha curcas] |
| 2 |
Hb_003058_230 |
0.1475093336 |
- |
- |
copper ion binding protein, putative [Ricinus communis] |
| 3 |
Hb_053709_040 |
0.1584224311 |
- |
- |
putative actin-97 protein [Linum usitatissimum] |
| 4 |
Hb_008185_020 |
0.1631508921 |
- |
- |
RING-H2 finger protein ATL3J, putative [Ricinus communis] |
| 5 |
Hb_000816_190 |
0.1674070866 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_007575_090 |
0.181967401 |
- |
- |
putative. proteolipid subunit of vacuolar H+ ATPase, partial [Zea mays] |
| 7 |
Hb_011918_010 |
0.1857376693 |
- |
- |
hypothetical protein glysoja_041948 [Glycine soja] |
| 8 |
Hb_034507_040 |
0.185872492 |
- |
- |
peptide transporter, putative [Ricinus communis] |
| 9 |
Hb_007632_100 |
0.1866232465 |
transcription factor |
TF Family: NF-YB |
ccaat-binding transcription factor subunit A, putative [Ricinus communis] |
| 10 |
Hb_007483_060 |
0.1878123221 |
desease resistance |
Gene Name: ABC_membrane |
multidrug resistance protein 1, 2, putative [Ricinus communis] |
| 11 |
Hb_002490_030 |
0.192418218 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000805_140 |
0.1936464723 |
transcription factor |
TF Family: GRAS |
Chitin-inducible gibberellin-responsive protein, putative [Ricinus communis] |
| 13 |
Hb_002900_100 |
0.1964378173 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase S.4-like [Jatropha curcas] |
| 14 |
Hb_184541_010 |
0.1998369672 |
- |
- |
PREDICTED: selT-like protein [Jatropha curcas] |
| 15 |
Hb_000311_040 |
0.2034531382 |
- |
- |
PREDICTED: S-formylglutathione hydrolase [Jatropha curcas] |
| 16 |
Hb_066514_010 |
0.2048880941 |
- |
- |
hypothetical protein RCOM_0204720 [Ricinus communis] |
| 17 |
Hb_003207_150 |
0.2069499459 |
- |
- |
PREDICTED: SPX domain-containing membrane protein At4g22990-like [Jatropha curcas] |
| 18 |
Hb_001486_050 |
0.2079054864 |
- |
- |
PREDICTED: arabinogalactan peptide 13-like [Jatropha curcas] |
| 19 |
Hb_000977_200 |
0.2102565034 |
- |
- |
PREDICTED: acyl-[acyl-carrier-protein] desaturase, chloroplastic-like [Jatropha curcas] |
| 20 |
Hb_002243_050 |
0.2103049565 |
- |
- |
PREDICTED: ion channel DMI1-like isoform X2 [Jatropha curcas] |