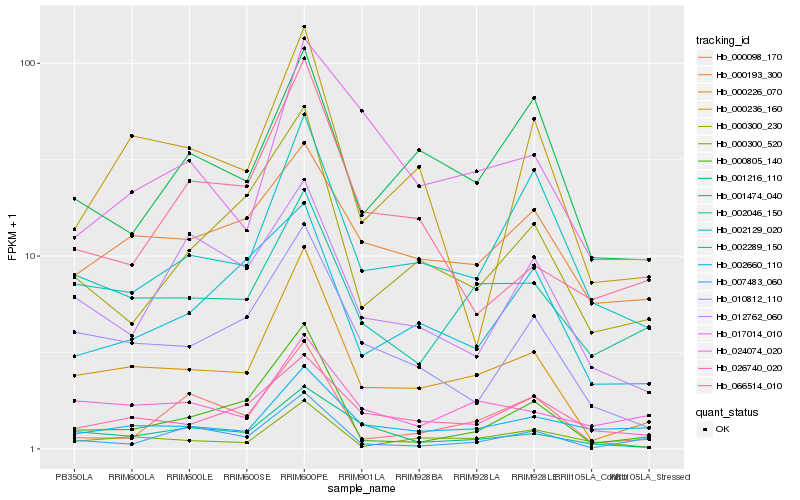
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000226_070 |
0.0 |
- |
- |
PREDICTED: pyrophosphate-energized vacuolar membrane proton pump [Jatropha curcas] |
| 2 |
Hb_010812_110 |
0.1473595824 |
- |
- |
putative plant disease resistance family protein [Populus trichocarpa] |
| 3 |
Hb_001474_040 |
0.1684895884 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 [Jatropha curcas] |
| 4 |
Hb_000300_230 |
0.1710299894 |
- |
- |
PREDICTED: cation/calcium exchanger 2 [Jatropha curcas] |
| 5 |
Hb_002129_020 |
0.1739984261 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX14 [Jatropha curcas] |
| 6 |
Hb_000300_520 |
0.1756975295 |
- |
- |
hypothetical protein JCGZ_02702 [Jatropha curcas] |
| 7 |
Hb_002046_150 |
0.1763144581 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g15080 [Jatropha curcas] |
| 8 |
Hb_026740_020 |
0.1794404372 |
desease resistance |
Gene Name: NB-ARC |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 9 |
Hb_000805_140 |
0.1797604195 |
transcription factor |
TF Family: GRAS |
Chitin-inducible gibberellin-responsive protein, putative [Ricinus communis] |
| 10 |
Hb_024074_020 |
0.1837527206 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g09600 [Jatropha curcas] |
| 11 |
Hb_000236_160 |
0.1850977203 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 6 [Jatropha curcas] |
| 12 |
Hb_001216_110 |
0.1882888766 |
- |
- |
PREDICTED: protein GPR107-like [Jatropha curcas] |
| 13 |
Hb_002660_110 |
0.1901274135 |
- |
- |
PREDICTED: serine/threonine-protein kinase-like protein CCR1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002289_150 |
0.1905348043 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 3 [UDP-forming]-like [Jatropha curcas] |
| 15 |
Hb_000193_300 |
0.1918747515 |
- |
- |
PREDICTED: putative glycosyltransferase 5 [Jatropha curcas] |
| 16 |
Hb_017014_010 |
0.1976984881 |
- |
- |
PREDICTED: syntaxin-132-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_007483_060 |
0.1978049312 |
desease resistance |
Gene Name: ABC_membrane |
multidrug resistance protein 1, 2, putative [Ricinus communis] |
| 18 |
Hb_000098_170 |
0.1980733556 |
- |
- |
hypothetical protein JCGZ_06491 [Jatropha curcas] |
| 19 |
Hb_012762_060 |
0.1994820334 |
- |
- |
Vitellogenic carboxypeptidase, putative [Ricinus communis] |
| 20 |
Hb_066514_010 |
0.1999865792 |
- |
- |
hypothetical protein RCOM_0204720 [Ricinus communis] |