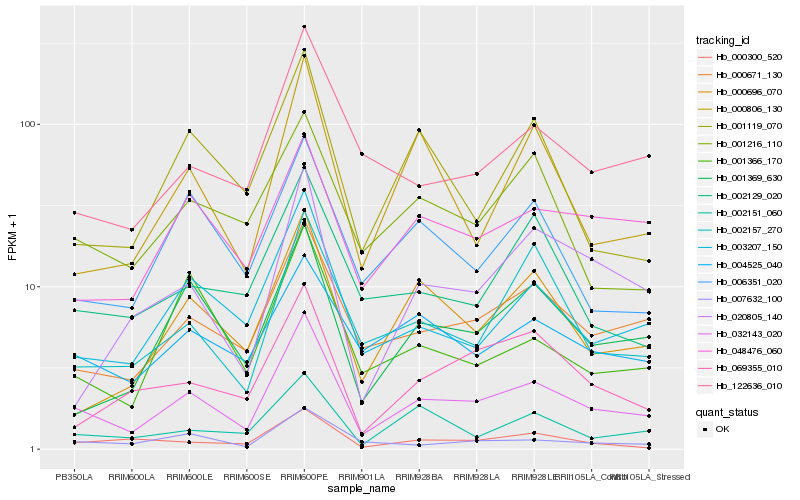
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_032143_020 |
0.0 |
- |
- |
hypothetical protein POPTR_0009s06310g [Populus trichocarpa] |
| 2 |
Hb_000671_130 |
0.1351235215 |
- |
- |
PREDICTED: CMP-sialic acid transporter 2 isoform X2 [Jatropha curcas] |
| 3 |
Hb_003207_150 |
0.1367424248 |
- |
- |
PREDICTED: SPX domain-containing membrane protein At4g22990-like [Jatropha curcas] |
| 4 |
Hb_001369_630 |
0.1581272783 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 3-like [Jatropha curcas] |
| 5 |
Hb_007632_100 |
0.1596131453 |
transcription factor |
TF Family: NF-YB |
ccaat-binding transcription factor subunit A, putative [Ricinus communis] |
| 6 |
Hb_122636_010 |
0.1644913413 |
- |
- |
dienelactone hydrolase family protein [Populus trichocarpa] |
| 7 |
Hb_000696_070 |
0.1673161715 |
- |
- |
PREDICTED: aspartic proteinase-like protein 1 [Jatropha curcas] |
| 8 |
Hb_048476_060 |
0.1678000114 |
- |
- |
PREDICTED: CRAL-TRIO domain-containing protein YKL091C-like isoform X2 [Jatropha curcas] |
| 9 |
Hb_004525_040 |
0.1684546473 |
- |
- |
PREDICTED: sugar transporter ERD6-like 6 [Jatropha curcas] |
| 10 |
Hb_006351_020 |
0.171242886 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase S.4-like [Jatropha curcas] |
| 11 |
Hb_002151_060 |
0.1727148568 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit rpc4 [Jatropha curcas] |
| 12 |
Hb_002157_270 |
0.1737978311 |
- |
- |
PREDICTED: transmembrane protein 87B [Jatropha curcas] |
| 13 |
Hb_000300_520 |
0.1744961115 |
- |
- |
hypothetical protein JCGZ_02702 [Jatropha curcas] |
| 14 |
Hb_000806_130 |
0.175447777 |
- |
- |
Adenosine kinase 2 [Theobroma cacao] |
| 15 |
Hb_001216_110 |
0.1769382505 |
- |
- |
PREDICTED: protein GPR107-like [Jatropha curcas] |
| 16 |
Hb_069355_010 |
0.1779063136 |
- |
- |
NBS-LRR resistance protein RGH2 [Manihot esculenta] |
| 17 |
Hb_001366_170 |
0.1783965069 |
- |
- |
PREDICTED: uncharacterized protein C594.04c [Jatropha curcas] |
| 18 |
Hb_002129_020 |
0.1790545395 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX14 [Jatropha curcas] |
| 19 |
Hb_001119_070 |
0.1798306959 |
- |
- |
methylenetetrahydrofolate reductase, putative [Ricinus communis] |
| 20 |
Hb_020805_140 |
0.1806495115 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At5g42360-like [Jatropha curcas] |