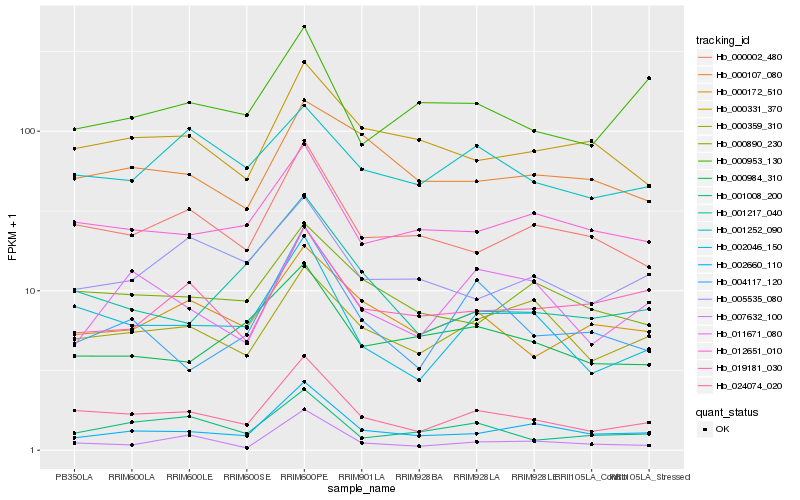
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_024074_020 |
0.0 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g09600 [Jatropha curcas] |
| 2 |
Hb_002046_150 |
0.0797942127 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g15080 [Jatropha curcas] |
| 3 |
Hb_000002_480 |
0.1238175269 |
- |
- |
PREDICTED: protein WVD2-like 1 isoform X2 [Jatropha curcas] |
| 4 |
Hb_002660_110 |
0.1265314926 |
- |
- |
PREDICTED: serine/threonine-protein kinase-like protein CCR1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001008_200 |
0.139018141 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC103961552 [Pyrus x bretschneideri] |
| 6 |
Hb_012651_010 |
0.1396057164 |
- |
- |
PREDICTED: uncharacterized protein LOC105645131 [Jatropha curcas] |
| 7 |
Hb_000172_510 |
0.1424041294 |
- |
- |
hypothetical protein B456_007G243100 [Gossypium raimondii] |
| 8 |
Hb_000359_310 |
0.143080935 |
- |
- |
hypothetical protein JCGZ_03934 [Jatropha curcas] |
| 9 |
Hb_000890_230 |
0.1471278843 |
- |
- |
hypothetical protein JCGZ_02013 [Jatropha curcas] |
| 10 |
Hb_001217_040 |
0.150959658 |
- |
- |
hypothetical protein MIMGU_mgv1a0002421mg, partial [Erythranthe guttata] |
| 11 |
Hb_000331_370 |
0.1515878157 |
- |
- |
Glyceraldehyde-3-phosphate dehydrogenase-2C cytosolic [Gossypium arboreum] |
| 12 |
Hb_004117_120 |
0.1518037572 |
- |
- |
Brassinosteroid LRR receptor kinase precursor, putative [Ricinus communis] |
| 13 |
Hb_007632_100 |
0.1545010211 |
transcription factor |
TF Family: NF-YB |
ccaat-binding transcription factor subunit A, putative [Ricinus communis] |
| 14 |
Hb_000984_310 |
0.1589640886 |
- |
- |
Hydrolase, hydrolyzing O-glycosyl compounds, putative [Theobroma cacao] |
| 15 |
Hb_005535_080 |
0.1599582878 |
- |
- |
hypothetical protein CICLE_v10012766mg [Citrus clementina] |
| 16 |
Hb_019181_030 |
0.1613419637 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: conserved oligomeric Golgi complex subunit 6-like [Pyrus x bretschneideri] |
| 17 |
Hb_001252_090 |
0.1614285374 |
- |
- |
GDP-D-mannose pyrophosphorylase [Camellia sinensis] |
| 18 |
Hb_000107_080 |
0.1634964725 |
- |
- |
PREDICTED: uncharacterized protein LOC105641009 [Jatropha curcas] |
| 19 |
Hb_011671_080 |
0.1637368262 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like protein kinase TDR [Jatropha curcas] |
| 20 |
Hb_000953_130 |
0.1642637866 |
- |
- |
PREDICTED: fructose-bisphosphate aldolase cytoplasmic isozyme [Jatropha curcas] |