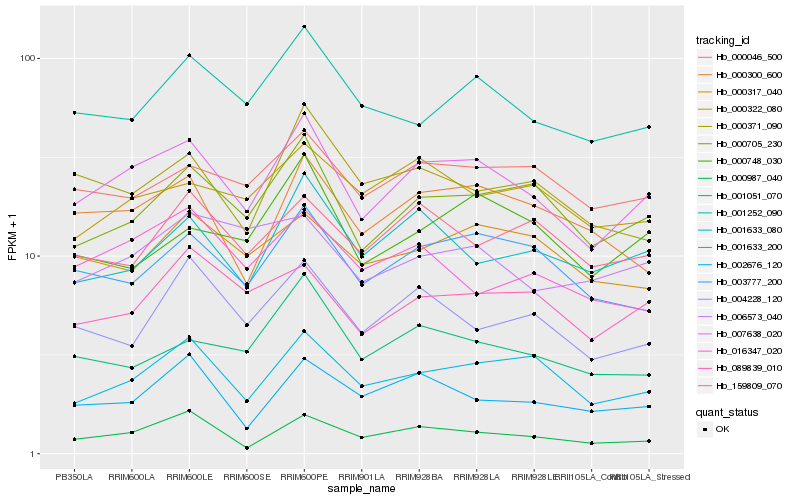
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007638_020 |
0.0 |
- |
- |
PREDICTED: low-temperature-induced cysteine proteinase-like [Jatropha curcas] |
| 2 |
Hb_000705_230 |
0.0907320362 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1 isozyme 2 [Jatropha curcas] |
| 3 |
Hb_001633_200 |
0.1019593701 |
- |
- |
hypothetical protein B456_005G137400 [Gossypium raimondii] |
| 4 |
Hb_003777_200 |
0.1034931041 |
- |
- |
PREDICTED: uncharacterized protein LOC105640933 [Jatropha curcas] |
| 5 |
Hb_001252_090 |
0.106928612 |
- |
- |
GDP-D-mannose pyrophosphorylase [Camellia sinensis] |
| 6 |
Hb_000046_500 |
0.111417843 |
- |
- |
PREDICTED: probable serine/threonine-protein phosphatase 2A regulatory subunit B'' subunit TON2 [Jatropha curcas] |
| 7 |
Hb_000300_600 |
0.1135379764 |
- |
- |
tubulin alpha chain, putative [Ricinus communis] |
| 8 |
Hb_089839_010 |
0.1136376786 |
- |
- |
PREDICTED: probable xyloglucan glycosyltransferase 6 [Jatropha curcas] |
| 9 |
Hb_001633_080 |
0.1157370993 |
- |
- |
PREDICTED: V-type proton ATPase subunit C [Gossypium raimondii] |
| 10 |
Hb_000987_040 |
0.1200050595 |
- |
- |
PREDICTED: protein BREAST CANCER SUSCEPTIBILITY 2 homolog B isoform X2 [Jatropha curcas] |
| 11 |
Hb_001051_070 |
0.1203443129 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000371_090 |
0.1210562259 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase 48 kDa subunit-like [Jatropha curcas] |
| 13 |
Hb_000317_040 |
0.121993283 |
- |
- |
PREDICTED: xylosyltransferase 1-like [Jatropha curcas] |
| 14 |
Hb_000748_030 |
0.1227140552 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 15 |
Hb_000322_080 |
0.1231141786 |
- |
- |
hypothetical protein JCGZ_07423 [Jatropha curcas] |
| 16 |
Hb_004228_120 |
0.1235422373 |
- |
- |
hypothetical protein POPTR_0013s02080g [Populus trichocarpa] |
| 17 |
Hb_016347_020 |
0.1246349872 |
- |
- |
PREDICTED: D-amino-acid transaminase, chloroplastic-like isoform X1 [Populus euphratica] |
| 18 |
Hb_159809_070 |
0.1252641476 |
- |
- |
phospholipase A-2-activating protein, putative [Ricinus communis] |
| 19 |
Hb_002676_120 |
0.1254902511 |
- |
- |
hypothetical protein POPTR_0002s23750g [Populus trichocarpa] |
| 20 |
Hb_006573_040 |
0.1257358111 |
- |
- |
DNA binding protein, putative [Ricinus communis] |