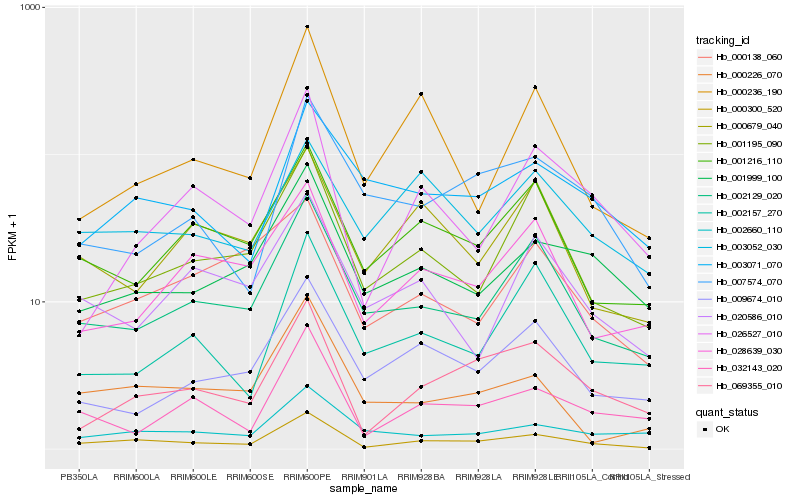
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000300_520 |
0.0 |
- |
- |
hypothetical protein JCGZ_02702 [Jatropha curcas] |
| 2 |
Hb_069355_010 |
0.151552172 |
- |
- |
NBS-LRR resistance protein RGH2 [Manihot esculenta] |
| 3 |
Hb_002129_020 |
0.1563211381 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX14 [Jatropha curcas] |
| 4 |
Hb_001195_090 |
0.1681731397 |
- |
- |
PREDICTED: isoflavone reductase-like protein [Jatropha curcas] |
| 5 |
Hb_001999_100 |
0.1708776035 |
- |
- |
PREDICTED: UDP-glycosyltransferase 71D1-like [Jatropha curcas] |
| 6 |
Hb_001216_110 |
0.1734337636 |
- |
- |
PREDICTED: protein GPR107-like [Jatropha curcas] |
| 7 |
Hb_032143_020 |
0.1744961115 |
- |
- |
hypothetical protein POPTR_0009s06310g [Populus trichocarpa] |
| 8 |
Hb_000226_070 |
0.1756975295 |
- |
- |
PREDICTED: pyrophosphate-energized vacuolar membrane proton pump [Jatropha curcas] |
| 9 |
Hb_028639_030 |
0.1921751579 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 2-like [Jatropha curcas] |
| 10 |
Hb_026527_010 |
0.1927514419 |
- |
- |
PREDICTED: caffeoylshikimate esterase-like [Populus euphratica] |
| 11 |
Hb_003071_070 |
0.192969564 |
- |
- |
sucrose synthase 3 [Hevea brasiliensis] |
| 12 |
Hb_000236_190 |
0.1933814838 |
- |
- |
PREDICTED: adenosylhomocysteinase 1 [Jatropha curcas] |
| 13 |
Hb_002660_110 |
0.1935444733 |
- |
- |
PREDICTED: serine/threonine-protein kinase-like protein CCR1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_009674_010 |
0.1947049554 |
- |
- |
AAEL007687-PA [Aedes aegypti] |
| 15 |
Hb_003052_030 |
0.1967836185 |
- |
- |
6-phosphogluconate dehydrogenase family protein isoform 1 [Theobroma cacao] |
| 16 |
Hb_000679_040 |
0.1971236948 |
- |
- |
Cellulose synthase 1 [Theobroma cacao] |
| 17 |
Hb_007574_070 |
0.1972040422 |
- |
- |
fatty acid desaturase, putative [Ricinus communis] |
| 18 |
Hb_000138_060 |
0.1985365611 |
- |
- |
PREDICTED: protein trichome birefringence [Jatropha curcas] |
| 19 |
Hb_020586_010 |
0.1995073213 |
- |
- |
Alpha-N-arabinofuranosidase 1 precursor, putative [Ricinus communis] |
| 20 |
Hb_002157_270 |
0.1996219947 |
- |
- |
PREDICTED: transmembrane protein 87B [Jatropha curcas] |