| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
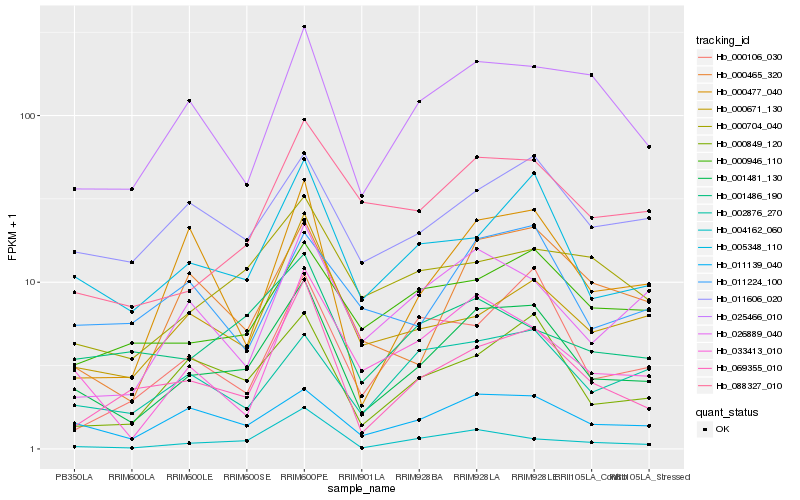
Hb_001481_130 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_005348_110 |
0.1380160408 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 3 |
Hb_011139_040 |
0.1422863636 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor SPATULA-like isoform X2 [Jatropha curcas] |
| 4 |
Hb_000849_120 |
0.1426536848 |
- |
- |
PREDICTED: linoleate 9S-lipoxygenase 6-like [Jatropha curcas] |
| 5 |
Hb_069355_010 |
0.1611322896 |
- |
- |
NBS-LRR resistance protein RGH2 [Manihot esculenta] |
| 6 |
Hb_000465_320 |
0.1650589494 |
- |
- |
PREDICTED: nudix hydrolase 8 [Jatropha curcas] |
| 7 |
Hb_033413_010 |
0.1669094161 |
- |
- |
Reticuline oxidase precursor, putative [Ricinus communis] |
| 8 |
Hb_011606_020 |
0.1677327336 |
- |
- |
hypothetical protein CISIN_1g023687mg [Citrus sinensis] |
| 9 |
Hb_025466_010 |
0.1692803888 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 10 |
Hb_088327_010 |
0.1698312986 |
- |
- |
PREDICTED: serine hydroxymethyltransferase 7 [Jatropha curcas] |
| 11 |
Hb_000946_110 |
0.1724808716 |
- |
- |
GTP cyclohydrolase I, putative [Ricinus communis] |
| 12 |
Hb_002876_270 |
0.1775903905 |
- |
- |
PREDICTED: probable magnesium transporter NIPA4 isoform X1 [Jatropha curcas] |
| 13 |
Hb_026889_040 |
0.1794491695 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Jatropha curcas] |
| 14 |
Hb_000477_040 |
0.1802767077 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_011224_100 |
0.1811305954 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000671_130 |
0.1838524056 |
- |
- |
PREDICTED: CMP-sialic acid transporter 2 isoform X2 [Jatropha curcas] |
| 17 |
Hb_004162_060 |
0.1842282609 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 18 |
Hb_000106_030 |
0.1867843829 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 19 |
Hb_000704_040 |
0.1868678627 |
- |
- |
aquaporin sip2.1, putative [Ricinus communis] |
| 20 |
Hb_001486_190 |
0.1880283635 |
- |
- |
hypothetical protein JCGZ_02702 [Jatropha curcas] |