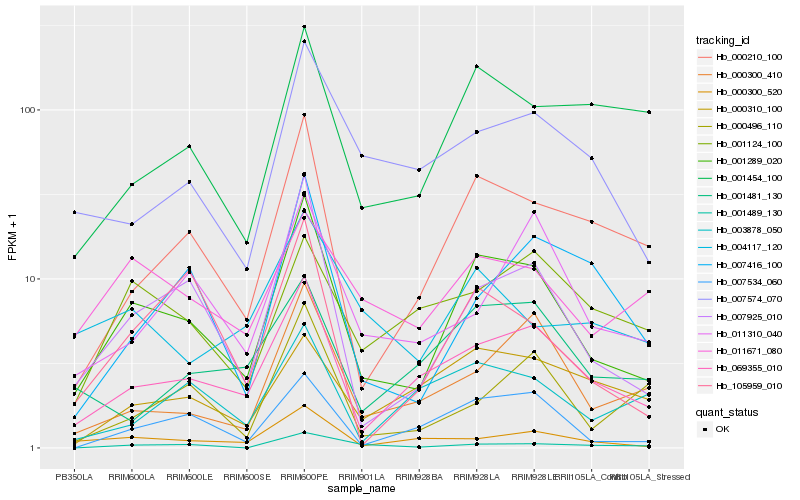
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001289_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105635827 [Jatropha curcas] |
| 2 |
Hb_007925_010 |
0.1505203613 |
- |
- |
PREDICTED: asparagine synthetase [glutamine-hydrolyzing] 1 [Jatropha curcas] |
| 3 |
Hb_069355_010 |
0.1629436093 |
- |
- |
NBS-LRR resistance protein RGH2 [Manihot esculenta] |
| 4 |
Hb_000210_100 |
0.1891048774 |
- |
- |
pepsin A, putative [Ricinus communis] |
| 5 |
Hb_007534_060 |
0.1918532675 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 6 |
Hb_000300_520 |
0.2090293483 |
- |
- |
hypothetical protein JCGZ_02702 [Jatropha curcas] |
| 7 |
Hb_105959_010 |
0.209150788 |
- |
- |
asparagine synthetase, putative [Ricinus communis] |
| 8 |
Hb_000300_410 |
0.2169347214 |
- |
- |
hypothetical protein POPTR_0013s05860g [Populus trichocarpa] |
| 9 |
Hb_007574_070 |
0.222586979 |
- |
- |
fatty acid desaturase, putative [Ricinus communis] |
| 10 |
Hb_001454_100 |
0.2291021101 |
- |
- |
PREDICTED: protein RALF-like 33 [Jatropha curcas] |
| 11 |
Hb_011671_080 |
0.2308357349 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like protein kinase TDR [Jatropha curcas] |
| 12 |
Hb_007416_100 |
0.2321914114 |
- |
- |
PREDICTED: probable xyloglucan glycosyltransferase 5 [Jatropha curcas] |
| 13 |
Hb_011310_040 |
0.2347169725 |
transcription factor |
TF Family: bZIP |
transcription factor hy5, putative [Ricinus communis] |
| 14 |
Hb_001489_130 |
0.2354635453 |
- |
- |
Pectate lyase precursor, putative [Ricinus communis] |
| 15 |
Hb_004117_120 |
0.2393961823 |
- |
- |
Brassinosteroid LRR receptor kinase precursor, putative [Ricinus communis] |
| 16 |
Hb_001481_130 |
0.2424675013 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000496_110 |
0.2427037582 |
- |
- |
PREDICTED: uncharacterized protein LOC105650214 [Jatropha curcas] |
| 18 |
Hb_003878_050 |
0.2441265781 |
- |
- |
PREDICTED: peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase A-like [Jatropha curcas] |
| 19 |
Hb_001124_100 |
0.2443671103 |
- |
- |
PREDICTED: vesicle-associated protein 2-2-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_000310_100 |
0.2481350308 |
- |
- |
conserved hypothetical protein [Ricinus communis] |