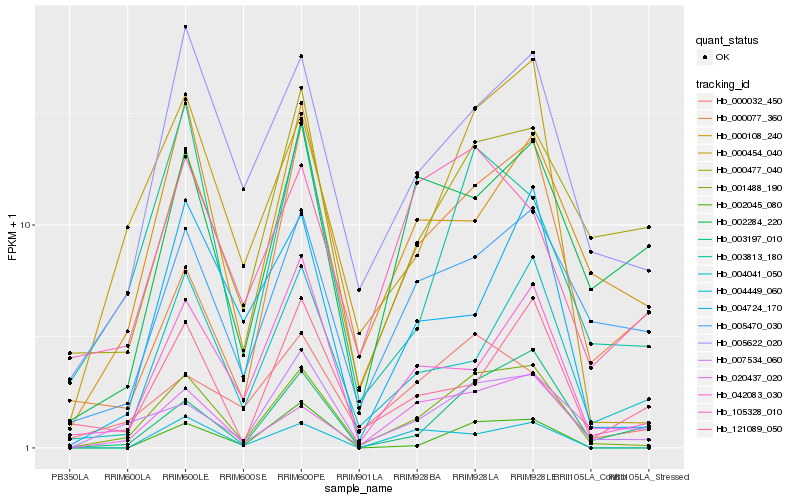
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001488_190 |
0.0 |
- |
- |
alpha-amylase, putative [Ricinus communis] |
| 2 |
Hb_000454_040 |
0.1453581507 |
- |
- |
PREDICTED: kinesin-like protein NACK1 [Jatropha curcas] |
| 3 |
Hb_007534_060 |
0.170010873 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 4 |
Hb_005622_020 |
0.1815822832 |
- |
- |
PREDICTED: DCC family protein At1g52590, chloroplastic [Jatropha curcas] |
| 5 |
Hb_003197_010 |
0.1981619203 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_003813_180 |
0.2054692184 |
- |
- |
sterol desaturase, putative [Ricinus communis] |
| 7 |
Hb_005470_030 |
0.2095417308 |
- |
- |
PREDICTED: probable galactinol--sucrose galactosyltransferase 2 isoform X2 [Jatropha curcas] |
| 8 |
Hb_004041_050 |
0.2102863252 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like [Jatropha curcas] |
| 9 |
Hb_042083_030 |
0.2121568672 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 10 |
Hb_002045_080 |
0.2135200405 |
- |
- |
PREDICTED: uncharacterized protein LOC105649907 [Jatropha curcas] |
| 11 |
Hb_000108_240 |
0.2166187977 |
- |
- |
hypothetical protein Csa_1G600150 [Cucumis sativus] |
| 12 |
Hb_004724_170 |
0.2202812447 |
- |
- |
PREDICTED: rho GTPase-activating protein 1 [Jatropha curcas] |
| 13 |
Hb_004449_060 |
0.2213318182 |
- |
- |
PREDICTED: uncharacterized protein LOC105631660 [Jatropha curcas] |
| 14 |
Hb_000032_450 |
0.2246261629 |
- |
- |
PREDICTED: GDSL esterase/lipase At1g71691 [Jatropha curcas] |
| 15 |
Hb_020437_020 |
0.2256914578 |
- |
- |
hypothetical protein POPTR_0001s01410g, partial [Populus trichocarpa] |
| 16 |
Hb_000477_040 |
0.2294062093 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_121089_050 |
0.230854771 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000077_360 |
0.2325818705 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 19 |
Hb_002284_220 |
0.232837095 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At5g42360-like [Jatropha curcas] |
| 20 |
Hb_105328_010 |
0.2354824981 |
- |
- |
PREDICTED: low-temperature-induced cysteine proteinase-like [Jatropha curcas] |