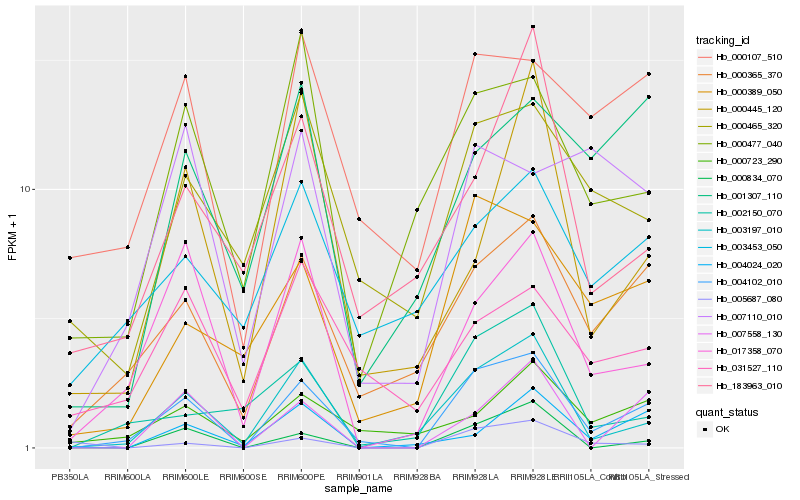
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004102_010 |
0.0 |
- |
- |
F-box family protein [Theobroma cacao] |
| 2 |
Hb_003197_010 |
0.1942115707 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000365_370 |
0.2128751259 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_017358_070 |
0.2340562795 |
- |
- |
snf1-kinase beta subunit, plants, putative [Ricinus communis] |
| 5 |
Hb_000834_070 |
0.2349890195 |
- |
- |
PREDICTED: tankyrase-1 isoform X4 [Jatropha curcas] |
| 6 |
Hb_000389_050 |
0.237005142 |
- |
- |
PREDICTED: uncharacterized protein LOC105650917 [Jatropha curcas] |
| 7 |
Hb_000107_510 |
0.2457048022 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 8 |
Hb_007558_130 |
0.2504679435 |
- |
- |
- |
| 9 |
Hb_000723_290 |
0.2576312132 |
- |
- |
calcium lipid binding protein, putative [Ricinus communis] |
| 10 |
Hb_002150_070 |
0.2607387351 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor CRF2-like [Jatropha curcas] |
| 11 |
Hb_000465_320 |
0.2656148258 |
- |
- |
PREDICTED: nudix hydrolase 8 [Jatropha curcas] |
| 12 |
Hb_000477_040 |
0.2686525884 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_004024_020 |
0.2766919923 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 14 |
Hb_001307_110 |
0.277630855 |
- |
- |
PREDICTED: uncharacterized protein LOC105644507 [Jatropha curcas] |
| 15 |
Hb_031527_110 |
0.2781183176 |
- |
- |
PREDICTED: probable glutathione S-transferase parC [Jatropha curcas] |
| 16 |
Hb_000445_120 |
0.2784727314 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 17 |
Hb_183963_010 |
0.2809358621 |
- |
- |
core region of GTP cyclohydrolase I family protein [Populus trichocarpa] |
| 18 |
Hb_003453_050 |
0.2822496327 |
- |
- |
PREDICTED: calcium-binding mitochondrial carrier protein SCaMC-3-like [Jatropha curcas] |
| 19 |
Hb_007110_010 |
0.2835933077 |
- |
- |
PREDICTED: accessory gland protein Acp36DE [Jatropha curcas] |
| 20 |
Hb_005687_080 |
0.2840579152 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |