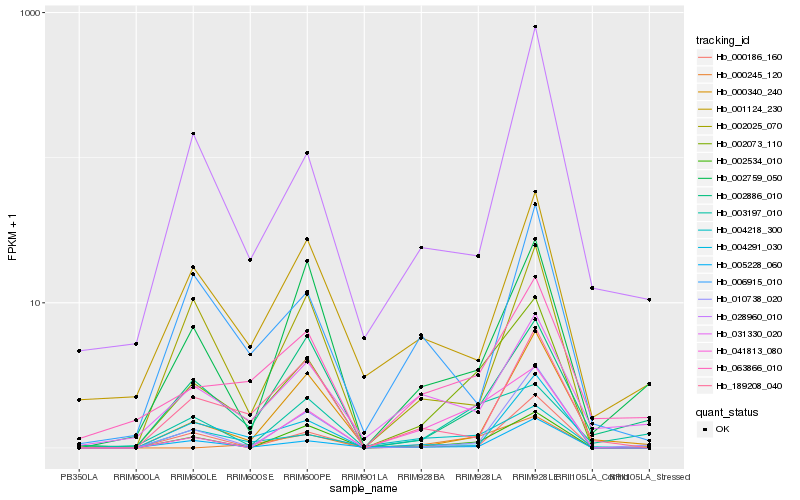
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002073_110 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002534_010 |
0.1884011553 |
- |
- |
PREDICTED: expansin-A1-like [Jatropha curcas] |
| 3 |
Hb_041813_080 |
0.1957570357 |
- |
- |
- |
| 4 |
Hb_004218_300 |
0.198523675 |
- |
- |
hypothetical protein JCGZ_24534 [Jatropha curcas] |
| 5 |
Hb_063866_010 |
0.2214914458 |
- |
- |
PREDICTED: uncharacterized protein LOC105632605 [Jatropha curcas] |
| 6 |
Hb_005228_060 |
0.2255425705 |
- |
- |
PREDICTED: uncharacterized protein LOC102604101 [Solanum tuberosum] |
| 7 |
Hb_004291_030 |
0.2271622397 |
- |
- |
PREDICTED: MATE efflux family protein 9-like [Jatropha curcas] |
| 8 |
Hb_002025_070 |
0.2287707701 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g13770, chloroplastic-like isoform X1 [Populus euphratica] |
| 9 |
Hb_000186_160 |
0.2351307069 |
- |
- |
hypothetical protein POPTR_0011s01120g [Populus trichocarpa] |
| 10 |
Hb_000245_120 |
0.2373987614 |
- |
- |
hypothetical protein POPTR_0001s18890g [Populus trichocarpa] |
| 11 |
Hb_010738_020 |
0.2377232346 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 12 |
Hb_002886_010 |
0.2430745032 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_189208_040 |
0.2431670843 |
- |
- |
hypothetical protein POPTR_0002s18920g [Populus trichocarpa] |
| 14 |
Hb_001124_230 |
0.2467446513 |
- |
- |
PREDICTED: divinyl chlorophyllide a 8-vinyl-reductase, chloroplastic [Jatropha curcas] |
| 15 |
Hb_002759_050 |
0.2485424442 |
- |
- |
PREDICTED: protein ALTERED XYLOGLUCAN 4 [Jatropha curcas] |
| 16 |
Hb_031330_020 |
0.2542703448 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like [Jatropha curcas] |
| 17 |
Hb_000340_240 |
0.255008148 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_003197_010 |
0.2552702538 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_006915_010 |
0.2557288675 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 20 |
Hb_028960_010 |
0.2570543275 |
- |
- |
phosphoglycerate kinase, putative [Ricinus communis] |