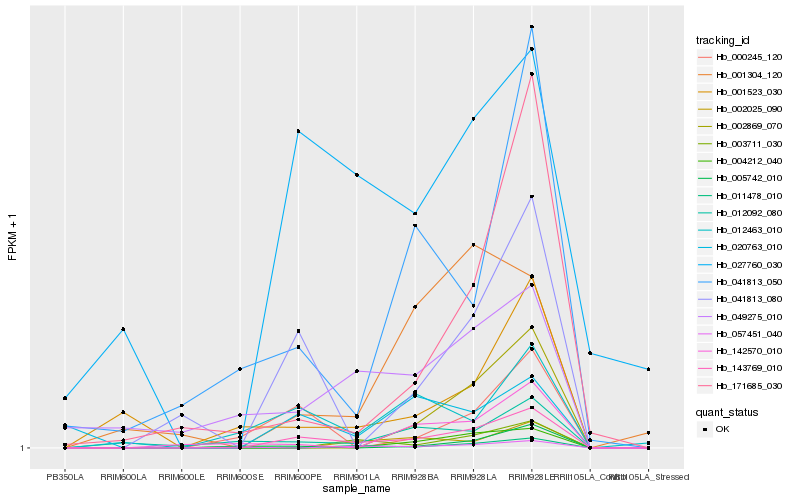
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_143769_010 |
0.0 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase MRH1 [Jatropha curcas] |
| 2 |
Hb_041813_080 |
0.2335897934 |
- |
- |
- |
| 3 |
Hb_000245_120 |
0.2703215204 |
- |
- |
hypothetical protein POPTR_0001s18890g [Populus trichocarpa] |
| 4 |
Hb_012463_010 |
0.2790465165 |
- |
- |
PREDICTED: uncharacterized protein LOC104901296 [Beta vulgaris subsp. vulgaris] |
| 5 |
Hb_011478_010 |
0.2829622059 |
- |
- |
- |
| 6 |
Hb_027760_030 |
0.2858438425 |
- |
- |
- |
| 7 |
Hb_020763_010 |
0.287410187 |
- |
- |
polyamine oxidase, putative [Ricinus communis] |
| 8 |
Hb_003711_030 |
0.2918785554 |
- |
- |
PREDICTED: uncharacterized protein LOC102621950 [Citrus sinensis] |
| 9 |
Hb_012092_080 |
0.2926486563 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g15280-like [Malus domestica] |
| 10 |
Hb_001523_030 |
0.2934874519 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 11 |
Hb_005742_010 |
0.2942508169 |
- |
- |
hypothetical protein, partial [Pseudomonas fluorescens] |
| 12 |
Hb_171685_030 |
0.3018430598 |
- |
- |
URF-1 [Citrullus lanatus] |
| 13 |
Hb_142570_010 |
0.3047007802 |
- |
- |
PREDICTED: uncharacterized protein LOC104907573 [Beta vulgaris subsp. vulgaris] |
| 14 |
Hb_049275_010 |
0.3057023093 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 4-like [Populus euphratica] |
| 15 |
Hb_041813_050 |
0.3066864679 |
- |
- |
hypothetical protein NitaMp096 [Nicotiana tabacum] |
| 16 |
Hb_001304_120 |
0.3086871453 |
transcription factor |
TF Family: WRKY |
WRKY protein [Hevea brasiliensis] |
| 17 |
Hb_002025_090 |
0.3089765775 |
- |
- |
unnamed protein product [Coffea canephora] |
| 18 |
Hb_057451_040 |
0.3098505507 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 19 |
Hb_004212_040 |
0.3158446905 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 20 |
Hb_002869_070 |
0.3173805597 |
- |
- |
- |