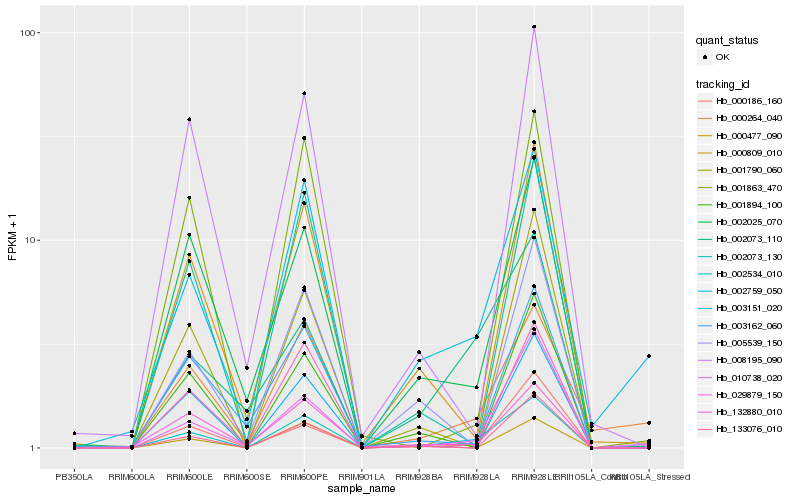
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002534_010 |
0.0 |
- |
- |
PREDICTED: expansin-A1-like [Jatropha curcas] |
| 2 |
Hb_002025_070 |
0.1647554067 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g13770, chloroplastic-like isoform X1 [Populus euphratica] |
| 3 |
Hb_002759_050 |
0.1664580494 |
- |
- |
PREDICTED: protein ALTERED XYLOGLUCAN 4 [Jatropha curcas] |
| 4 |
Hb_029879_150 |
0.1704416583 |
desease resistance |
Gene Name: NB-ARC |
putative disease resistance gene NBS-LRR family protein [Populus trichocarpa] |
| 5 |
Hb_002073_130 |
0.1847530699 |
- |
- |
PREDICTED: uncharacterized protein LOC100246830 [Vitis vinifera] |
| 6 |
Hb_000477_090 |
0.1871590913 |
- |
- |
PREDICTED: uncharacterized protein LOC105639302 [Jatropha curcas] |
| 7 |
Hb_002073_110 |
0.1884011553 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_010738_020 |
0.191782885 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 9 |
Hb_000186_160 |
0.194884785 |
- |
- |
hypothetical protein POPTR_0011s01120g [Populus trichocarpa] |
| 10 |
Hb_001894_100 |
0.1964069003 |
- |
- |
PREDICTED: kininogen-1-like [Jatropha curcas] |
| 11 |
Hb_133076_010 |
0.1982420036 |
- |
- |
PREDICTED: scopoletin glucosyltransferase-like [Populus euphratica] |
| 12 |
Hb_001863_470 |
0.1982765882 |
- |
- |
PREDICTED: uncharacterized protein LOC105630613 [Jatropha curcas] |
| 13 |
Hb_001790_060 |
0.1990281603 |
- |
- |
PREDICTED: uncharacterized protein LOC105644961 [Jatropha curcas] |
| 14 |
Hb_000809_010 |
0.2004380274 |
- |
- |
hypothetical protein JCGZ_12342 [Jatropha curcas] |
| 15 |
Hb_000264_040 |
0.2017206914 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 16 |
Hb_003151_020 |
0.2021465204 |
- |
- |
Ankyrin repeat family protein, putative [Theobroma cacao] |
| 17 |
Hb_132880_010 |
0.2027539309 |
- |
- |
PREDICTED: uncharacterized protein LOC105641595 [Jatropha curcas] |
| 18 |
Hb_003162_060 |
0.2040449272 |
- |
- |
PREDICTED: MATH domain-containing protein At5g43560-like [Jatropha curcas] |
| 19 |
Hb_005539_150 |
0.2041051435 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 33 [Jatropha curcas] |
| 20 |
Hb_008195_090 |
0.2043938558 |
- |
- |
conserved hypothetical protein [Ricinus communis] |