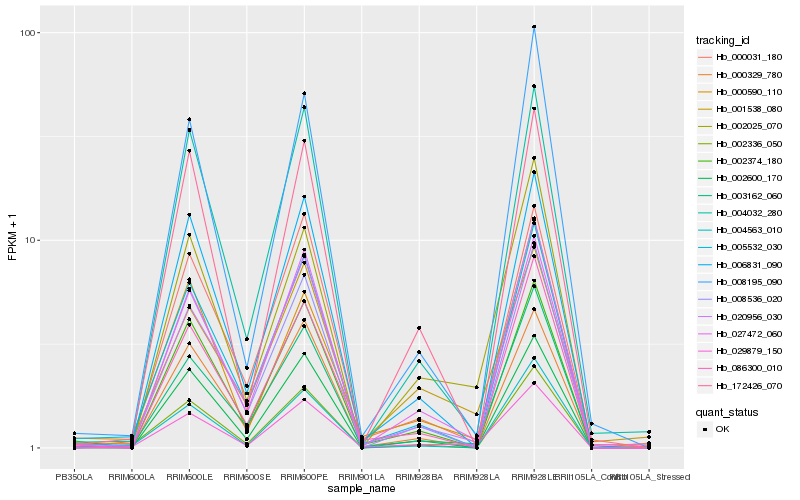
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_029879_150 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
putative disease resistance gene NBS-LRR family protein [Populus trichocarpa] |
| 2 |
Hb_003162_060 |
0.1004459829 |
- |
- |
PREDICTED: MATH domain-containing protein At5g43560-like [Jatropha curcas] |
| 3 |
Hb_002025_070 |
0.1081595593 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g13770, chloroplastic-like isoform X1 [Populus euphratica] |
| 4 |
Hb_020956_030 |
0.1092222191 |
- |
- |
PREDICTED: swi5-dependent recombination DNA repair protein 1 homolog [Jatropha curcas] |
| 5 |
Hb_002336_050 |
0.1100597259 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 6 |
Hb_027472_060 |
0.111629509 |
transcription factor |
TF Family: GRAS |
DELLA protein GAI1, putative [Ricinus communis] |
| 7 |
Hb_005532_030 |
0.1162228733 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 8 |
Hb_006831_090 |
0.119762174 |
- |
- |
PREDICTED: uncharacterized protein LOC105642125 isoform X1 [Jatropha curcas] |
| 9 |
Hb_004032_280 |
0.1209523484 |
- |
- |
PREDICTED: uncharacterized protein LOC105635149 [Jatropha curcas] |
| 10 |
Hb_000031_180 |
0.1209864601 |
- |
- |
sarcosine oxidase, putative [Ricinus communis] |
| 11 |
Hb_004563_010 |
0.1213497834 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 12 |
Hb_086300_010 |
0.1222167877 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 13 |
Hb_001538_080 |
0.1249492574 |
- |
- |
PREDICTED: QWRF motif-containing protein 2 isoform X2 [Jatropha curcas] |
| 14 |
Hb_008195_090 |
0.1258593052 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000590_110 |
0.1272660601 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_008536_020 |
0.1297827816 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RHA2B [Jatropha curcas] |
| 17 |
Hb_002600_170 |
0.1305315569 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_172426_070 |
0.1306734738 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 3-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_000329_780 |
0.1322062215 |
- |
- |
hypothetical protein POPTR_0403s00200g [Populus trichocarpa] |
| 20 |
Hb_002374_180 |
0.1326463058 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER-related [Jatropha curcas] |