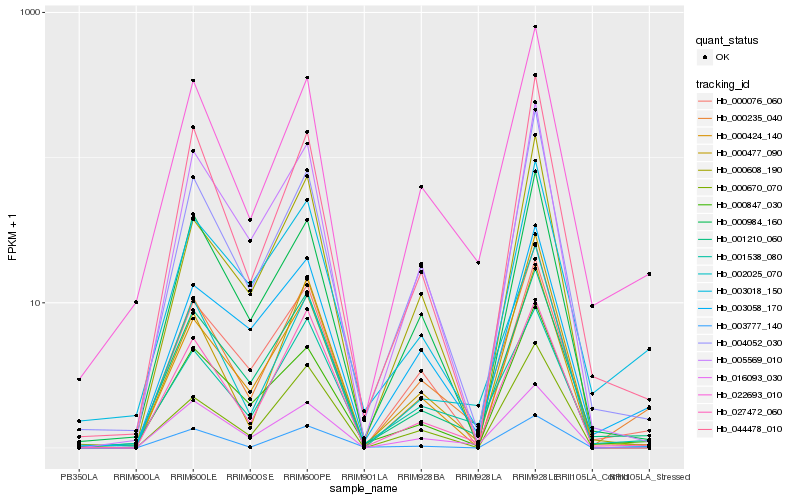
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000235_040 |
0.0 |
- |
- |
PREDICTED: probable receptor-like serine/threonine-protein kinase At5g57670 isoform X1 [Populus euphratica] |
| 2 |
Hb_022693_010 |
0.0892056272 |
- |
- |
pectin acetylesterase, putative [Ricinus communis] |
| 3 |
Hb_001210_060 |
0.1089395188 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 1-like [Jatropha curcas] |
| 4 |
Hb_002025_070 |
0.1129961814 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g13770, chloroplastic-like isoform X1 [Populus euphratica] |
| 5 |
Hb_000076_060 |
0.1137758241 |
- |
- |
hypothetical protein RCOM_0586940 [Ricinus communis] |
| 6 |
Hb_027472_060 |
0.1149689717 |
transcription factor |
TF Family: GRAS |
DELLA protein GAI1, putative [Ricinus communis] |
| 7 |
Hb_001538_080 |
0.1180937479 |
- |
- |
PREDICTED: QWRF motif-containing protein 2 isoform X2 [Jatropha curcas] |
| 8 |
Hb_004052_030 |
0.1197599723 |
- |
- |
PREDICTED: uncharacterized protein LOC105640637 [Jatropha curcas] |
| 9 |
Hb_000424_140 |
0.1201009664 |
- |
- |
Protein kinase APK1A, chloroplast precursor, putative [Ricinus communis] |
| 10 |
Hb_003058_170 |
0.1220235182 |
- |
- |
PREDICTED: F-box protein At2g26850 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000847_030 |
0.1258625982 |
- |
- |
PREDICTED: asparagine--tRNA ligase, cytoplasmic 2 [Jatropha curcas] |
| 12 |
Hb_000477_090 |
0.125923948 |
- |
- |
PREDICTED: uncharacterized protein LOC105639302 [Jatropha curcas] |
| 13 |
Hb_003777_140 |
0.1263682277 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_003018_150 |
0.1264234859 |
- |
- |
PREDICTED: uncharacterized protein LOC105628869 [Jatropha curcas] |
| 15 |
Hb_005569_010 |
0.1271322642 |
- |
- |
Heme-binding protein, putative [Ricinus communis] |
| 16 |
Hb_044478_010 |
0.1275980114 |
- |
- |
plastoquinol-plastocyanin reductase, putative [Ricinus communis] |
| 17 |
Hb_000984_160 |
0.1279286766 |
- |
- |
PREDICTED: amino acid permease 6-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_000670_070 |
0.1289858174 |
- |
- |
PREDICTED: probable carbohydrate esterase At4g34215 [Jatropha curcas] |
| 19 |
Hb_016093_030 |
0.1294180689 |
- |
- |
ankyrin repeat protein [Bruguiera gymnorhiza] |
| 20 |
Hb_000608_190 |
0.134537969 |
rubber biosynthesis |
Gene Name: Isopentenyl-diphosphate delta isomerase |
PREDICTED: isopentenyl-diphosphate Delta-isomerase I [Jatropha curcas] |