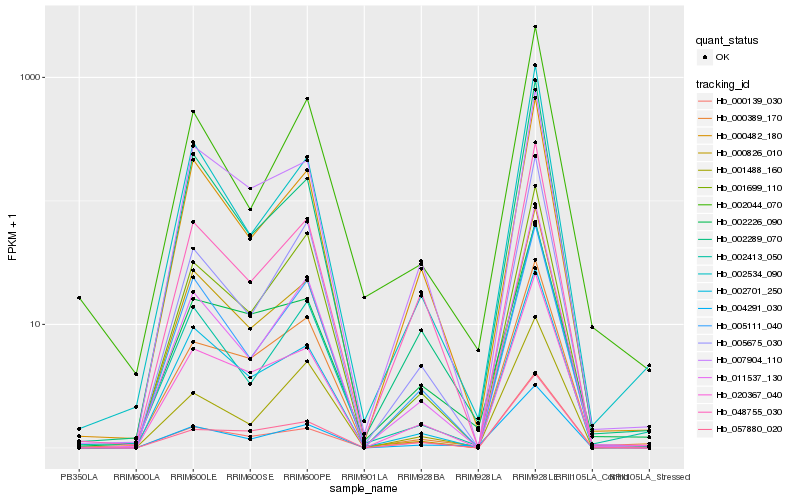
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004291_030 |
0.0 |
- |
- |
PREDICTED: MATE efflux family protein 9-like [Jatropha curcas] |
| 2 |
Hb_000482_180 |
0.0957177873 |
- |
- |
PREDICTED: glycine dehydrogenase (decarboxylating), mitochondrial [Jatropha curcas] |
| 3 |
Hb_011537_130 |
0.0980776239 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 4 |
Hb_048755_030 |
0.0989670825 |
- |
- |
Periplasmic beta-glucosidase precursor, putative [Ricinus communis] |
| 5 |
Hb_020367_040 |
0.1004925258 |
- |
- |
hypothetical protein JCGZ_01061 [Jatropha curcas] |
| 6 |
Hb_005675_030 |
0.10369168 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 6 [Jatropha curcas] |
| 7 |
Hb_002701_250 |
0.1063417753 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |
| 8 |
Hb_002226_090 |
0.1093639332 |
- |
- |
Stachyose synthase precursor, putative [Ricinus communis] |
| 9 |
Hb_002534_090 |
0.1103770444 |
- |
- |
geranylgeranyl reductase [Hevea brasiliensis] |
| 10 |
Hb_002289_070 |
0.1120124607 |
- |
- |
PREDICTED: magnesium-chelatase subunit ChlH, chloroplastic [Jatropha curcas] |
| 11 |
Hb_001488_160 |
0.1165228525 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 12 |
Hb_001699_110 |
0.1167220535 |
- |
- |
PREDICTED: pathogenesis-related protein 5-like [Jatropha curcas] |
| 13 |
Hb_057880_020 |
0.1182185406 |
- |
- |
PREDICTED: receptor-like protein kinase HSL1, partial [Jatropha curcas] |
| 14 |
Hb_000826_010 |
0.1185021353 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_005111_040 |
0.118931006 |
- |
- |
glucose-6-phosphate dehydrogenase [Hevea brasiliensis] |
| 16 |
Hb_002413_050 |
0.12080519 |
- |
- |
hypothetical protein POPTR_0018s06150g [Populus trichocarpa] |
| 17 |
Hb_000389_170 |
0.120919342 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4-like [Jatropha curcas] |
| 18 |
Hb_002044_070 |
0.1228015282 |
- |
- |
Chlorophyll a-b binding protein 3, chloroplastic [Theobroma cacao] |
| 19 |
Hb_007904_110 |
0.1232074599 |
- |
- |
PREDICTED: photosystem II core complex proteins psbY, chloroplastic-like [Populus euphratica] |
| 20 |
Hb_000139_030 |
0.1245862658 |
- |
- |
PREDICTED: uncharacterized protein LOC101254423 [Solanum lycopersicum] |