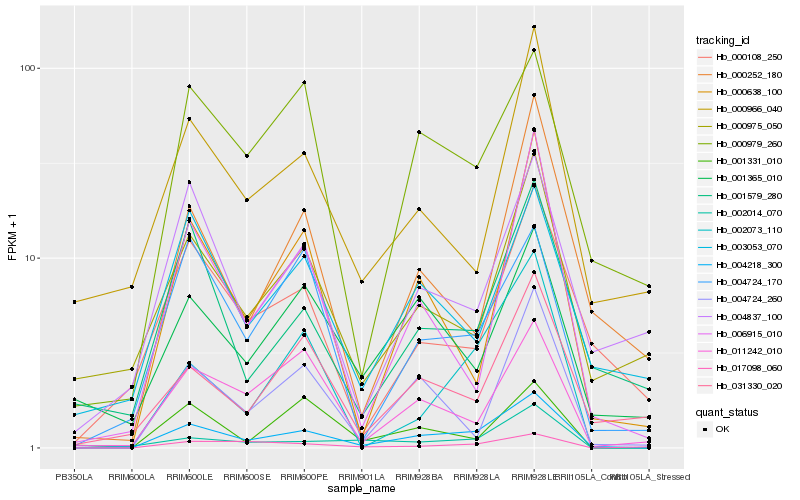
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004218_300 |
0.0 |
- |
- |
hypothetical protein JCGZ_24534 [Jatropha curcas] |
| 2 |
Hb_002073_110 |
0.198523675 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000638_100 |
0.2006365801 |
- |
- |
PREDICTED: histone deacetylase 14 [Jatropha curcas] |
| 4 |
Hb_004724_260 |
0.2055211736 |
- |
- |
PREDICTED: 40S ribosomal protein S14-2-like [Gossypium raimondii] |
| 5 |
Hb_006915_010 |
0.2064539703 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 6 |
Hb_031330_020 |
0.2089070412 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like [Jatropha curcas] |
| 7 |
Hb_002014_070 |
0.210563238 |
- |
- |
PREDICTED: probable chlorophyll(ide) b reductase NYC1, chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_000979_260 |
0.2109103786 |
- |
- |
ARF GTPase activator, putative [Ricinus communis] |
| 9 |
Hb_000252_180 |
0.2127103999 |
- |
- |
Glutamate--glyoxylate aminotransferase 2 [Morus notabilis] |
| 10 |
Hb_001579_280 |
0.2143430376 |
- |
- |
PREDICTED: 1,4-dihydroxy-2-naphthoyl-CoA synthase, peroxisomal-like [Jatropha curcas] |
| 11 |
Hb_011242_010 |
0.2181207393 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-2 [Jatropha curcas] |
| 12 |
Hb_000975_050 |
0.2188575274 |
- |
- |
PREDICTED: inner membrane protein PPF-1, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000108_250 |
0.2198605109 |
- |
- |
PREDICTED: protochlorophyllide-dependent translocon component 52, chloroplastic-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_003053_070 |
0.2216621408 |
- |
- |
PREDICTED: phosphoglucan phosphatase LSF1, chloroplastic [Jatropha curcas] |
| 15 |
Hb_017098_060 |
0.2249490943 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Pyrus x bretschneideri] |
| 16 |
Hb_004837_100 |
0.2250247291 |
- |
- |
PREDICTED: dicarboxylate transporter 1, chloroplastic [Jatropha curcas] |
| 17 |
Hb_001365_010 |
0.2251920787 |
- |
- |
hypothetical protein JCGZ_02762 [Jatropha curcas] |
| 18 |
Hb_001331_010 |
0.2255010956 |
- |
- |
hypothetical protein CISIN_1g039303mg, partial [Citrus sinensis] |
| 19 |
Hb_000966_040 |
0.2261524892 |
- |
- |
hypothetical protein JCGZ_01642 [Jatropha curcas] |
| 20 |
Hb_004724_170 |
0.2265051366 |
- |
- |
PREDICTED: rho GTPase-activating protein 1 [Jatropha curcas] |