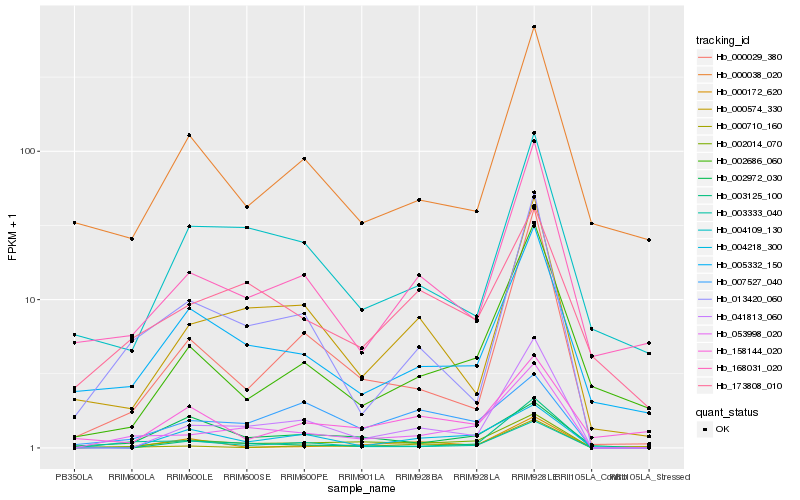
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002014_070 |
0.0 |
- |
- |
PREDICTED: probable chlorophyll(ide) b reductase NYC1, chloroplastic isoform X1 [Jatropha curcas] |
| 2 |
Hb_053998_020 |
0.1462826487 |
- |
- |
- |
| 3 |
Hb_041813_060 |
0.2001096154 |
- |
- |
ribosomal protein L10 (mitochondrion) [Ricinus communis] |
| 4 |
Hb_004218_300 |
0.210563238 |
- |
- |
hypothetical protein JCGZ_24534 [Jatropha curcas] |
| 5 |
Hb_002972_030 |
0.2182138889 |
- |
- |
protein with unknown function [Ricinus communis] |
| 6 |
Hb_005332_150 |
0.2198419285 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g46610 [Jatropha curcas] |
| 7 |
Hb_000029_380 |
0.2214261559 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000574_330 |
0.2276666111 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 9 |
Hb_007527_040 |
0.2282767877 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 2 [Jatropha curcas] |
| 10 |
Hb_003333_040 |
0.2306309796 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 11 |
Hb_002686_060 |
0.2324834301 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000172_620 |
0.2333615449 |
- |
- |
hypothetical protein VITISV_033646 [Vitis vinifera] |
| 13 |
Hb_168031_020 |
0.24042268 |
- |
- |
PREDICTED: uncharacterized protein LOC105646684 isoform X2 [Jatropha curcas] |
| 14 |
Hb_004109_130 |
0.2407899211 |
- |
- |
PREDICTED: translation initiation factor IF-2, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000038_020 |
0.245226062 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 2, chloroplastic [Jatropha curcas] |
| 16 |
Hb_003125_100 |
0.2457655522 |
- |
- |
NAD(P)-binding Rossmann-fold superfamily protein [Theobroma cacao] |
| 17 |
Hb_173808_010 |
0.2458366697 |
- |
- |
PREDICTED: translocase of chloroplast 159, chloroplastic [Jatropha curcas] |
| 18 |
Hb_013420_060 |
0.2466649018 |
- |
- |
NADH dehydrogenase subunit 7 [Nymphaea sp. Bergthorsson 0401] |
| 19 |
Hb_000710_160 |
0.2469955751 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 20 |
Hb_158144_020 |
0.2477019469 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Vitis vinifera] |