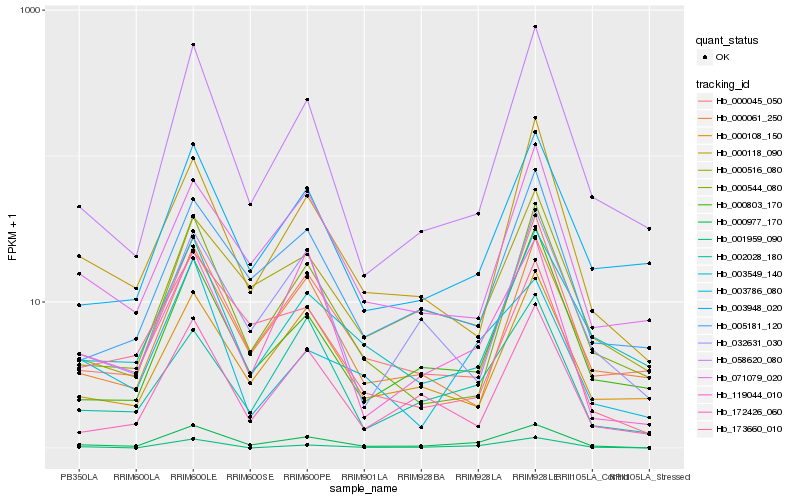
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000977_170 |
0.0 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 2 |
Hb_119044_010 |
0.157501892 |
- |
- |
PREDICTED: stomatal closure-related actin-binding protein 2-like [Jatropha curcas] |
| 3 |
Hb_000045_050 |
0.1596740273 |
- |
- |
PREDICTED: probable zinc metalloprotease EGY2, chloroplastic [Jatropha curcas] |
| 4 |
Hb_058620_080 |
0.159776552 |
- |
- |
PREDICTED: phosphoglycerate kinase, chloroplastic [Jatropha curcas] |
| 5 |
Hb_003948_020 |
0.1679354723 |
- |
- |
PREDICTED: transmembrane protein 41B [Jatropha curcas] |
| 6 |
Hb_000516_080 |
0.1685733248 |
- |
- |
hypothetical protein POPTR_0010s24810g [Populus trichocarpa] |
| 7 |
Hb_005181_120 |
0.171000172 |
- |
- |
PREDICTED: psbB mRNA maturation factor Mbb1, chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_002028_180 |
0.1717602189 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 9 |
Hb_000544_080 |
0.1730680687 |
- |
- |
PREDICTED: uncharacterized protein LOC105631062 isoform X3 [Jatropha curcas] |
| 10 |
Hb_000803_170 |
0.1734387849 |
- |
- |
PREDICTED: uncharacterized protein LOC105648312 [Jatropha curcas] |
| 11 |
Hb_172426_060 |
0.1740507966 |
- |
- |
PREDICTED: protein BREAST CANCER SUSCEPTIBILITY 1 homolog [Jatropha curcas] |
| 12 |
Hb_173660_010 |
0.1747172255 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Malus domestica] |
| 13 |
Hb_000108_150 |
0.1757065183 |
- |
- |
alpha/beta hydrolase, putative [Ricinus communis] |
| 14 |
Hb_000061_250 |
0.1760891287 |
- |
- |
hypothetical protein JCGZ_11665 [Jatropha curcas] |
| 15 |
Hb_003549_140 |
0.1761699457 |
- |
- |
Thylakoid lumenal 21.5 kDa protein, chloroplast precursor, putative [Ricinus communis] |
| 16 |
Hb_001959_090 |
0.1762910048 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Jatropha curcas] |
| 17 |
Hb_000118_090 |
0.1764083849 |
- |
- |
Glutamyl-tRNA reductase 1, chloroplast precursor, putative [Ricinus communis] |
| 18 |
Hb_071079_020 |
0.1768024494 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 19 |
Hb_003786_080 |
0.1772589881 |
- |
- |
electron transporter, putative [Ricinus communis] |
| 20 |
Hb_032631_030 |
0.1774808035 |
- |
- |
phosphatidic acid phosphatase, putative [Ricinus communis] |