| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
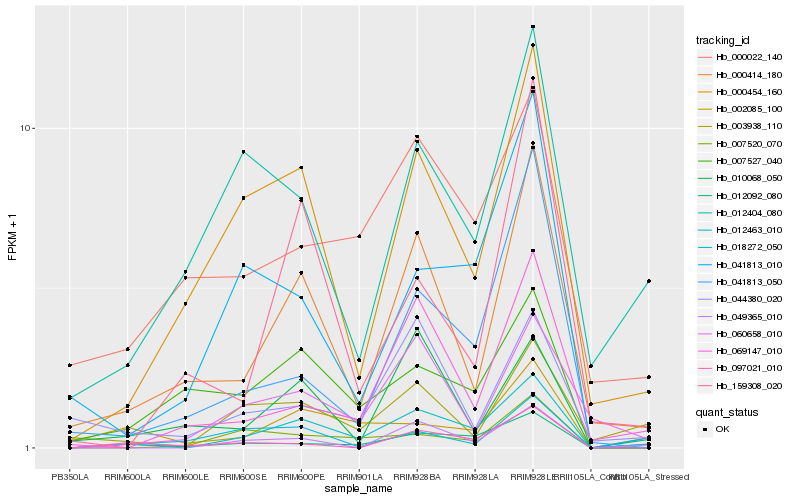
Hb_012463_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104901296 [Beta vulgaris subsp. vulgaris] |
| 2 |
Hb_012092_080 |
0.2047749923 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g15280-like [Malus domestica] |
| 3 |
Hb_000414_180 |
0.2145896478 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g27290 isoform X2 [Vitis vinifera] |
| 4 |
Hb_049365_010 |
0.2148016988 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Eucalyptus grandis] |
| 5 |
Hb_000454_160 |
0.2167113448 |
- |
- |
amidophosphoribosyltransferase, putative [Ricinus communis] |
| 6 |
Hb_003938_110 |
0.2400044742 |
- |
- |
polyprotein [Ananas comosus] |
| 7 |
Hb_069147_010 |
0.2400090011 |
desease resistance |
Gene Name: zf-BED |
PREDICTED: probable disease resistance protein At4g27220 isoform X1 [Populus euphratica] |
| 8 |
Hb_041813_010 |
0.24067601 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_041813_050 |
0.2413313659 |
- |
- |
hypothetical protein NitaMp096 [Nicotiana tabacum] |
| 10 |
Hb_002085_100 |
0.2443706177 |
- |
- |
PREDICTED: syntaxin-72 isoform X1 [Jatropha curcas] |
| 11 |
Hb_060658_010 |
0.2468243908 |
desease resistance |
Gene Name: NB-ARC |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 12 |
Hb_007520_070 |
0.2492500659 |
- |
- |
PREDICTED: uncharacterized protein LOC105155919 [Sesamum indicum] |
| 13 |
Hb_007527_040 |
0.2606112539 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 2 [Jatropha curcas] |
| 14 |
Hb_000022_140 |
0.2634516183 |
- |
- |
hypothetical protein MIMGU_mgv1a009920mg [Erythranthe guttata] |
| 15 |
Hb_097021_010 |
0.2657862962 |
- |
- |
hypothetical protein POPTR_0075s00200g, partial [Populus trichocarpa] |
| 16 |
Hb_012404_080 |
0.2661632535 |
- |
- |
PREDICTED: ABC transporter G family member 34-like [Jatropha curcas] |
| 17 |
Hb_018272_050 |
0.2687309816 |
- |
- |
hypothetical protein VITISV_037409 [Vitis vinifera] |
| 18 |
Hb_159308_020 |
0.2689511185 |
- |
- |
hypothetical protein B456_002G027900 [Gossypium raimondii] |
| 19 |
Hb_010068_050 |
0.2690695501 |
- |
- |
PREDICTED: uncharacterized protein LOC105632963 [Jatropha curcas] |
| 20 |
Hb_044380_020 |
0.2705118319 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |