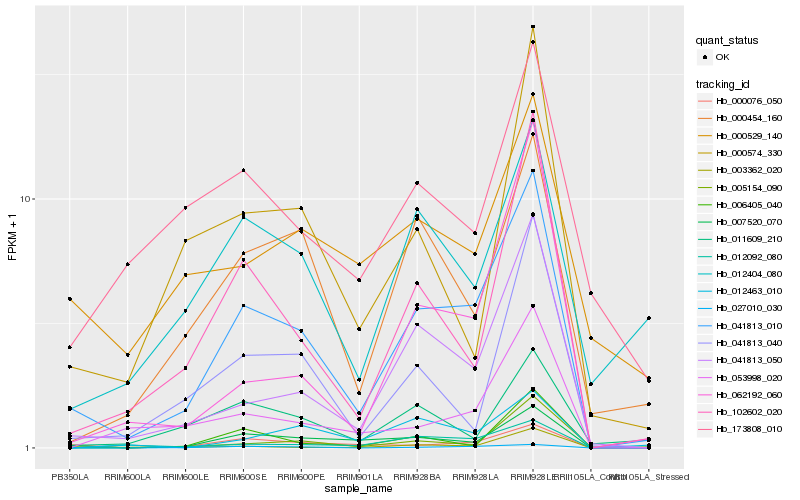
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_041813_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_041813_050 |
0.1744355866 |
- |
- |
hypothetical protein NitaMp096 [Nicotiana tabacum] |
| 3 |
Hb_102602_020 |
0.183725709 |
- |
- |
hypothetical protein B456_001G166300 [Gossypium raimondii] |
| 4 |
Hb_000076_050 |
0.191633771 |
- |
- |
PREDICTED: uncharacterized protein LOC105155919 [Sesamum indicum] |
| 5 |
Hb_007520_070 |
0.1983034568 |
- |
- |
PREDICTED: uncharacterized protein LOC105155919 [Sesamum indicum] |
| 6 |
Hb_000454_160 |
0.2075539611 |
- |
- |
amidophosphoribosyltransferase, putative [Ricinus communis] |
| 7 |
Hb_027010_030 |
0.2085801168 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 8 |
Hb_012092_080 |
0.2141299339 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g15280-like [Malus domestica] |
| 9 |
Hb_006405_040 |
0.2223568694 |
- |
- |
hypothetical protein VITISV_026680 [Vitis vinifera] |
| 10 |
Hb_041813_040 |
0.2224968396 |
- |
- |
NADH2 dehydrogenase (ubiquinone) (EC 1.6.5.3) chain 5, truncated - sugar beet mitochondrion (mitochondrion) [Beta vulgaris subsp. vulgaris] |
| 11 |
Hb_012404_080 |
0.222659555 |
- |
- |
PREDICTED: ABC transporter G family member 34-like [Jatropha curcas] |
| 12 |
Hb_003362_020 |
0.2233151004 |
- |
- |
PREDICTED: uncharacterized protein LOC104098435, partial [Nicotiana tomentosiformis] |
| 13 |
Hb_053998_020 |
0.2253986288 |
- |
- |
- |
| 14 |
Hb_062192_060 |
0.2285347348 |
- |
- |
maturase (mitochondrion) [Hevea brasiliensis] |
| 15 |
Hb_000574_330 |
0.229685346 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 16 |
Hb_005154_090 |
0.2329027136 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 17 |
Hb_011609_210 |
0.2342030502 |
- |
- |
Uncharacterized protein TCM_011393 [Theobroma cacao] |
| 18 |
Hb_012463_010 |
0.24067601 |
- |
- |
PREDICTED: uncharacterized protein LOC104901296 [Beta vulgaris subsp. vulgaris] |
| 19 |
Hb_000529_140 |
0.2473983427 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_173808_010 |
0.247915323 |
- |
- |
PREDICTED: translocase of chloroplast 159, chloroplastic [Jatropha curcas] |