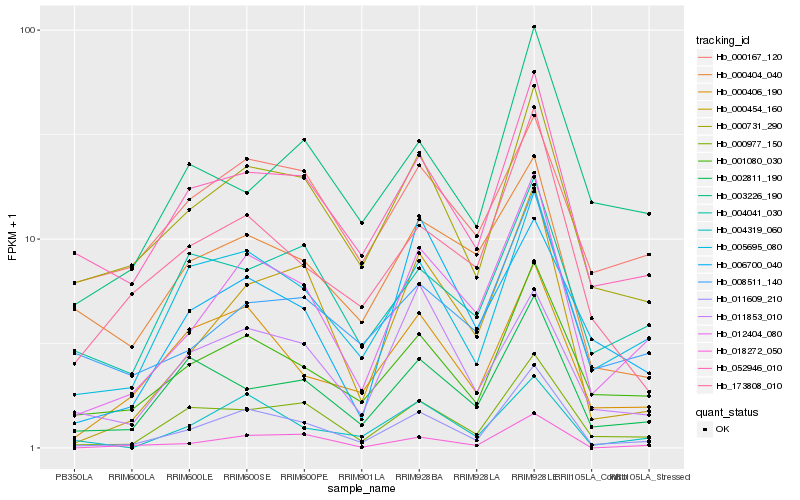
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012404_080 |
0.0 |
- |
- |
PREDICTED: ABC transporter G family member 34-like [Jatropha curcas] |
| 2 |
Hb_011609_210 |
0.1141729016 |
- |
- |
Uncharacterized protein TCM_011393 [Theobroma cacao] |
| 3 |
Hb_000454_160 |
0.133531999 |
- |
- |
amidophosphoribosyltransferase, putative [Ricinus communis] |
| 4 |
Hb_001080_030 |
0.1395101567 |
- |
- |
AMP dependent ligase, putative [Ricinus communis] |
| 5 |
Hb_000977_150 |
0.1494400633 |
- |
- |
latex cyanogenic beta glucosidase [Hevea brasiliensis] |
| 6 |
Hb_000731_290 |
0.1571897281 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 7 |
Hb_005695_080 |
0.1574286518 |
- |
- |
PREDICTED: acetyl-CoA carboxylase 1-like [Jatropha curcas] |
| 8 |
Hb_008511_140 |
0.1575530741 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105632314 [Jatropha curcas] |
| 9 |
Hb_052946_010 |
0.1583549506 |
- |
- |
PREDICTED: uncharacterized protein LOC105639103 [Jatropha curcas] |
| 10 |
Hb_004319_060 |
0.1736819094 |
- |
- |
PREDICTED: uncharacterized protein LOC104106381 [Nicotiana tomentosiformis] |
| 11 |
Hb_000404_040 |
0.1740869246 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002811_190 |
0.1741311352 |
- |
- |
tRNA pseudouridine synthase d, putative [Ricinus communis] |
| 13 |
Hb_018272_050 |
0.1742114895 |
- |
- |
hypothetical protein VITISV_037409 [Vitis vinifera] |
| 14 |
Hb_173808_010 |
0.1761907596 |
- |
- |
PREDICTED: translocase of chloroplast 159, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000406_190 |
0.1775643089 |
desease resistance |
Gene Name: NB-ARC |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 16 |
Hb_006700_040 |
0.1816931373 |
- |
- |
PREDICTED: vacuolar-sorting receptor 6-like [Jatropha curcas] |
| 17 |
Hb_004041_030 |
0.1843403661 |
- |
- |
PREDICTED: mitochondrial substrate carrier family protein C-like isoform X1 [Populus euphratica] |
| 18 |
Hb_003226_190 |
0.1882622547 |
- |
- |
PREDICTED: blue-light photoreceptor PHR2 [Jatropha curcas] |
| 19 |
Hb_011853_010 |
0.188302652 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 [Jatropha curcas] |
| 20 |
Hb_000167_120 |
0.1883334309 |
- |
- |
PREDICTED: U-box domain-containing protein 14 [Jatropha curcas] |