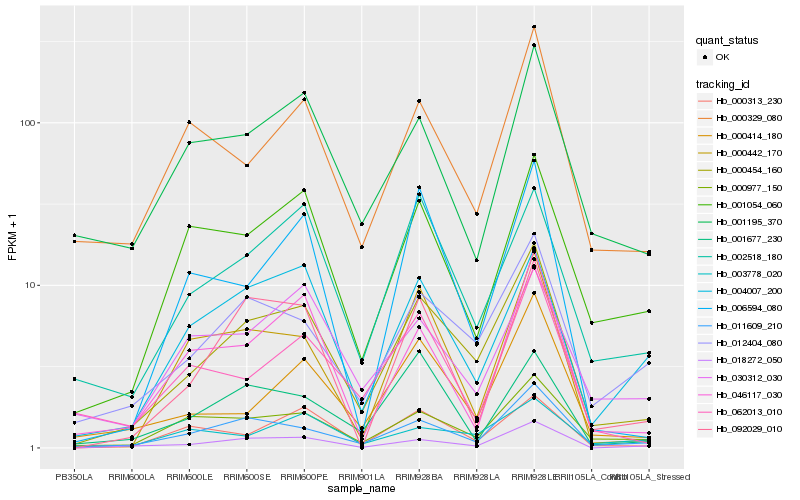
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000454_160 |
0.0 |
- |
- |
amidophosphoribosyltransferase, putative [Ricinus communis] |
| 2 |
Hb_012404_080 |
0.133531999 |
- |
- |
PREDICTED: ABC transporter G family member 34-like [Jatropha curcas] |
| 3 |
Hb_011609_210 |
0.1351383039 |
- |
- |
Uncharacterized protein TCM_011393 [Theobroma cacao] |
| 4 |
Hb_018272_050 |
0.1466753738 |
- |
- |
hypothetical protein VITISV_037409 [Vitis vinifera] |
| 5 |
Hb_000977_150 |
0.157969162 |
- |
- |
latex cyanogenic beta glucosidase [Hevea brasiliensis] |
| 6 |
Hb_002518_180 |
0.162571109 |
- |
- |
hypothetical protein POPTR_0001s25460g [Populus trichocarpa] |
| 7 |
Hb_000414_180 |
0.165018837 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g27290 isoform X2 [Vitis vinifera] |
| 8 |
Hb_004007_200 |
0.1746492957 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein NPY5 [Jatropha curcas] |
| 9 |
Hb_000313_230 |
0.1767804303 |
- |
- |
PREDICTED: uncharacterized protein LOC105636893 [Jatropha curcas] |
| 10 |
Hb_001054_060 |
0.17697812 |
- |
- |
PREDICTED: dihydropyrimidine dehydrogenase [NADP(+)] [Jatropha curcas] |
| 11 |
Hb_092029_010 |
0.1781560882 |
- |
- |
Disease resistance protein RPM1, putative [Ricinus communis] |
| 12 |
Hb_000329_080 |
0.179026733 |
- |
- |
PREDICTED: transketolase, chloroplastic [Jatropha curcas] |
| 13 |
Hb_046117_030 |
0.1817220364 |
- |
- |
sialyltransferase, putative [Ricinus communis] |
| 14 |
Hb_000442_170 |
0.1852982406 |
- |
- |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 15 |
Hb_001195_370 |
0.1859231888 |
- |
- |
- |
| 16 |
Hb_006594_080 |
0.1863195686 |
- |
- |
beta-glucosidase, putative [Ricinus communis] |
| 17 |
Hb_062013_010 |
0.1866540761 |
- |
- |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 18 |
Hb_003778_020 |
0.1886771397 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 19 |
Hb_030312_030 |
0.1938163692 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP40 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001677_230 |
0.1948553172 |
desease resistance |
Gene Name: NB-ARC |
conserved hypothetical protein [Ricinus communis] |