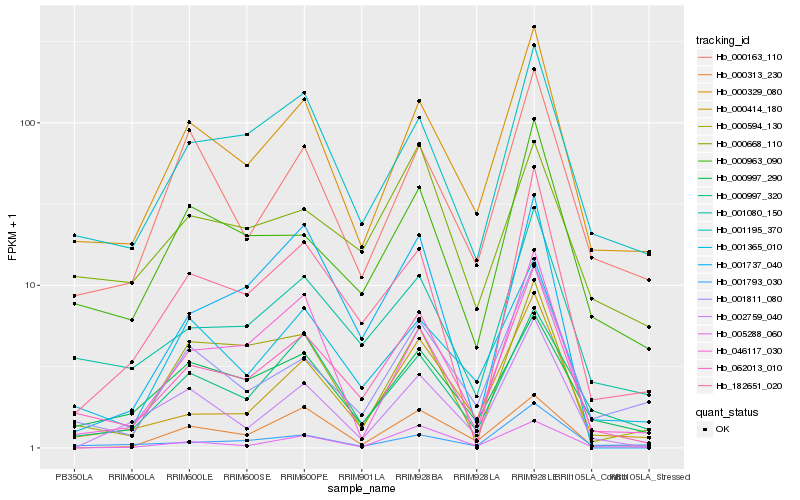
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_062013_010 |
0.0 |
- |
- |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 2 |
Hb_000329_080 |
0.1182965354 |
- |
- |
PREDICTED: transketolase, chloroplastic [Jatropha curcas] |
| 3 |
Hb_001080_150 |
0.1188127602 |
- |
- |
PREDICTED: Niemann-Pick C1 protein-like [Jatropha curcas] |
| 4 |
Hb_000414_180 |
0.123816606 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g27290 isoform X2 [Vitis vinifera] |
| 5 |
Hb_000963_090 |
0.1363226762 |
rubber biosynthesis |
Gene Name: 4-hydroxy-3-methylbut-2-enyl diphosphate reductase |
4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase [Hevea brasiliensis] |
| 6 |
Hb_001365_010 |
0.1445940696 |
- |
- |
hypothetical protein JCGZ_02762 [Jatropha curcas] |
| 7 |
Hb_182651_020 |
0.1461192762 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 8 |
Hb_001195_370 |
0.1499996414 |
- |
- |
- |
| 9 |
Hb_001811_080 |
0.1524612444 |
- |
- |
PREDICTED: fructose-1,6-bisphosphatase, cytosolic [Jatropha curcas] |
| 10 |
Hb_000997_290 |
0.1621976436 |
- |
- |
PREDICTED: acylamino-acid-releasing enzyme [Jatropha curcas] |
| 11 |
Hb_000997_320 |
0.16673765 |
- |
- |
PREDICTED: acylamino-acid-releasing enzyme-like [Malus domestica] |
| 12 |
Hb_000594_130 |
0.1681621107 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 13 |
Hb_001737_040 |
0.1736799467 |
- |
- |
PREDICTED: uncharacterized protein LOC104100193 [Nicotiana tomentosiformis] |
| 14 |
Hb_000163_110 |
0.1744346571 |
- |
- |
PREDICTED: blue-light photoreceptor PHR2 [Jatropha curcas] |
| 15 |
Hb_001793_030 |
0.1764105232 |
- |
- |
Cysteine-rich RLK (RECEPTOR-like protein kinase) 8 [Theobroma cacao] |
| 16 |
Hb_002759_040 |
0.1781386419 |
- |
- |
BnaA09g29830D [Brassica napus] |
| 17 |
Hb_005288_060 |
0.1784294266 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein At4g27190-like isoform X1 [Citrus sinensis] |
| 18 |
Hb_046117_030 |
0.1787929826 |
- |
- |
sialyltransferase, putative [Ricinus communis] |
| 19 |
Hb_000313_230 |
0.1796888976 |
- |
- |
PREDICTED: uncharacterized protein LOC105636893 [Jatropha curcas] |
| 20 |
Hb_000668_110 |
0.1799802186 |
- |
- |
PREDICTED: acetyl-CoA carboxylase 1-like [Jatropha curcas] |