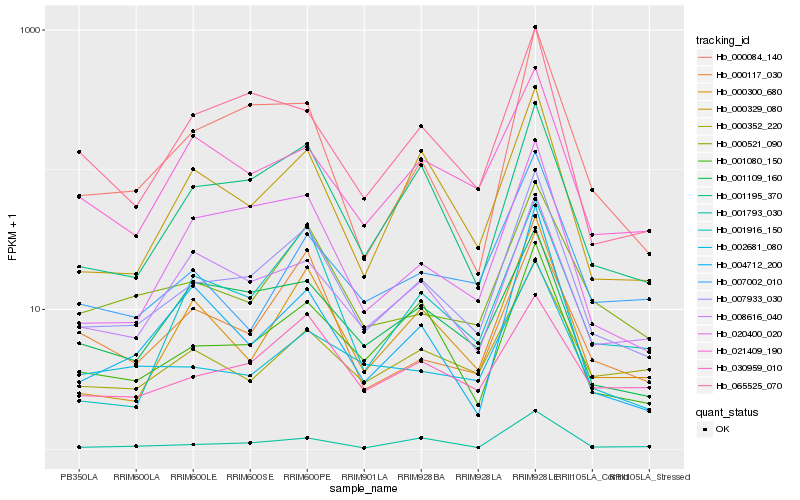
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007933_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105639932 [Jatropha curcas] |
| 2 |
Hb_000521_090 |
0.1202658936 |
- |
- |
PREDICTED: RING-H2 finger protein ATL5 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001195_370 |
0.1227674635 |
- |
- |
- |
| 4 |
Hb_001080_150 |
0.1237741318 |
- |
- |
PREDICTED: Niemann-Pick C1 protein-like [Jatropha curcas] |
| 5 |
Hb_001793_030 |
0.1275699429 |
- |
- |
Cysteine-rich RLK (RECEPTOR-like protein kinase) 8 [Theobroma cacao] |
| 6 |
Hb_000300_680 |
0.1277406166 |
- |
- |
PREDICTED: uncharacterized protein LOC105632625 isoform X1 [Jatropha curcas] |
| 7 |
Hb_020400_020 |
0.1287405983 |
- |
- |
starch synthase isoform II [Manihot esculenta] |
| 8 |
Hb_000084_140 |
0.1300819716 |
- |
- |
Aspartic proteinase nepenthesin-2 precursor, putative [Ricinus communis] |
| 9 |
Hb_002681_080 |
0.1363991001 |
- |
- |
PREDICTED: uncharacterized protein LOC105633817 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000329_080 |
0.1374828422 |
- |
- |
PREDICTED: transketolase, chloroplastic [Jatropha curcas] |
| 11 |
Hb_008616_040 |
0.1391929897 |
- |
- |
PREDICTED: RNA-binding protein CP33, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000352_220 |
0.1457712035 |
- |
- |
PREDICTED: probable tyrosine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 13 |
Hb_021409_190 |
0.1460807688 |
- |
- |
PREDICTED: luminal-binding protein 5-like [Jatropha curcas] |
| 14 |
Hb_000117_030 |
0.1494853298 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 [Jatropha curcas] |
| 15 |
Hb_065525_070 |
0.1501827864 |
- |
- |
unnamed protein product [Homo sapiens] |
| 16 |
Hb_030959_010 |
0.1513358484 |
- |
- |
cullin, putative [Ricinus communis] |
| 17 |
Hb_001109_160 |
0.1514356932 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 18 |
Hb_001916_150 |
0.1523805009 |
transcription factor |
TF Family: NF-YB |
PREDICTED: nuclear transcription factor Y subunit B-3 [Vitis vinifera] |
| 19 |
Hb_007002_010 |
0.1526184307 |
- |
- |
PREDICTED: dnaJ protein ERDJ3B [Jatropha curcas] |
| 20 |
Hb_004712_200 |
0.153837257 |
- |
- |
hypothetical protein CICLE_v10010292mg [Citrus clementina] |