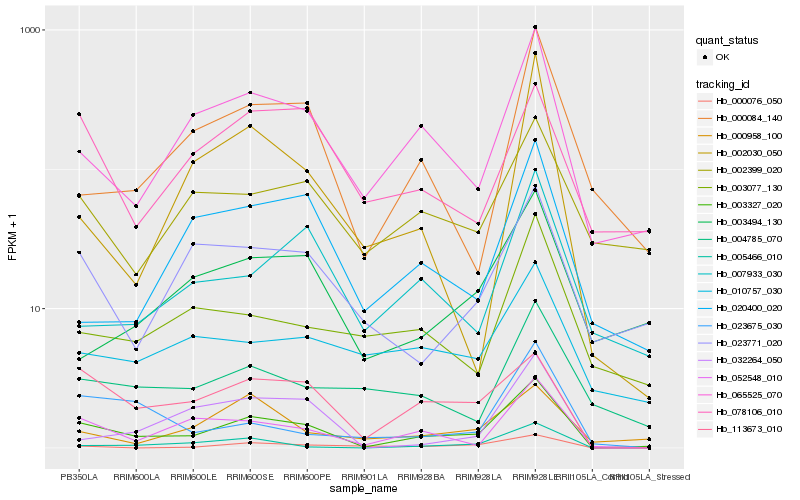
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003327_020 |
0.0 |
- |
- |
PREDICTED: phytosulfokine receptor 2-like, partial [Jatropha curcas] |
| 2 |
Hb_065525_070 |
0.1960957555 |
- |
- |
unnamed protein product [Homo sapiens] |
| 3 |
Hb_113673_010 |
0.2023667122 |
- |
- |
hypothetical protein POPTR_1763s00200g, partial [Populus trichocarpa] |
| 4 |
Hb_052548_010 |
0.2051776154 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 5 |
Hb_005466_010 |
0.2209066415 |
- |
- |
ATPase subunit 6, partial (mitochondrion) [Celastrus scandens] |
| 6 |
Hb_000084_140 |
0.2392419943 |
- |
- |
Aspartic proteinase nepenthesin-2 precursor, putative [Ricinus communis] |
| 7 |
Hb_023771_020 |
0.2435737766 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 isoform X3 [Jatropha curcas] |
| 8 |
Hb_032264_050 |
0.2481246444 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26 [Jatropha curcas] |
| 9 |
Hb_000076_050 |
0.2486510256 |
- |
- |
PREDICTED: uncharacterized protein LOC105155919 [Sesamum indicum] |
| 10 |
Hb_004785_070 |
0.2498054028 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_003077_130 |
0.2500561232 |
- |
- |
hypothetical protein JCGZ_22476 [Jatropha curcas] |
| 12 |
Hb_002399_020 |
0.2511595259 |
- |
- |
PREDICTED: luminal-binding protein 5-like [Jatropha curcas] |
| 13 |
Hb_002030_050 |
0.2511738483 |
- |
- |
classical arabinogalactan protein 7 precursor [Jatropha curcas] |
| 14 |
Hb_007933_030 |
0.2533836485 |
- |
- |
PREDICTED: uncharacterized protein LOC105639932 [Jatropha curcas] |
| 15 |
Hb_003494_130 |
0.253781829 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 9-like [Jatropha curcas] |
| 16 |
Hb_000958_100 |
0.2538541825 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 17 |
Hb_010757_030 |
0.2540054882 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 18 |
Hb_023675_030 |
0.2565301807 |
- |
- |
- |
| 19 |
Hb_020400_020 |
0.2577242803 |
- |
- |
starch synthase isoform II [Manihot esculenta] |
| 20 |
Hb_078106_010 |
0.2582824527 |
- |
- |
seed maturation protein PM37 [Populus trichocarpa] |