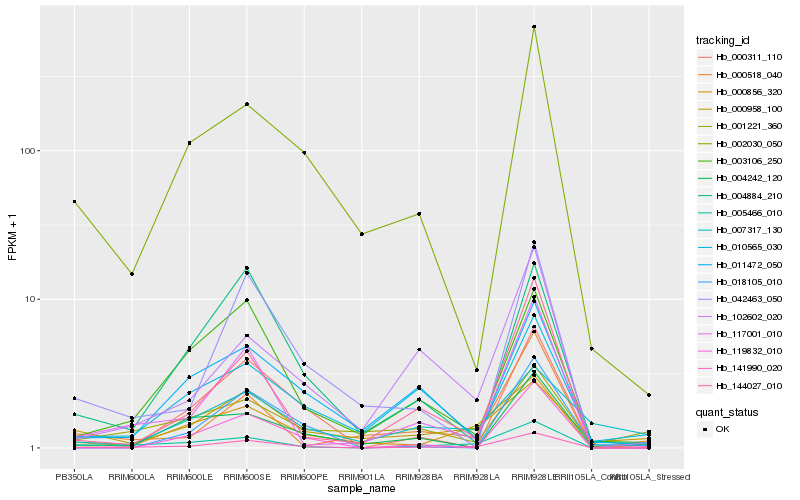
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_141990_020 |
0.0 |
- |
- |
RecName: Full=Genome polyprotein; Contains: RecName: Full=Movement protein; Short=MP; Contains: RecName: Full=Capsid protein; Short=CP; Contains: RecName: Full=Aspartic protease; Short=PR; Contains: RecName: Full=Reverse transcriptase; Short=RT [Petunia vein clearing virus isolate Hohn] |
| 2 |
Hb_000856_320 |
0.1813754389 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin of 36 kDa-like [Jatropha curcas] |
| 3 |
Hb_005466_010 |
0.2073806968 |
- |
- |
ATPase subunit 6, partial (mitochondrion) [Celastrus scandens] |
| 4 |
Hb_117001_010 |
0.2201478232 |
- |
- |
- |
| 5 |
Hb_010565_030 |
0.2291769394 |
- |
- |
hypothetical protein VITISV_006810 [Vitis vinifera] |
| 6 |
Hb_001221_360 |
0.2309294972 |
- |
- |
late embryogenesis abundant, putative [Ricinus communis] |
| 7 |
Hb_003106_250 |
0.232674694 |
- |
- |
hypothetical protein POPTR_0008s13520g [Populus trichocarpa] |
| 8 |
Hb_102602_020 |
0.2395883827 |
- |
- |
hypothetical protein B456_001G166300 [Gossypium raimondii] |
| 9 |
Hb_000518_040 |
0.2409107581 |
- |
- |
- |
| 10 |
Hb_000958_100 |
0.2418943042 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 11 |
Hb_144027_010 |
0.2428283989 |
- |
- |
- |
| 12 |
Hb_011472_050 |
0.2452281389 |
- |
- |
- |
| 13 |
Hb_004884_210 |
0.2457302768 |
- |
- |
PREDICTED: mitoferrin-like [Jatropha curcas] |
| 14 |
Hb_119832_010 |
0.2493389277 |
- |
- |
- |
| 15 |
Hb_000311_110 |
0.2501197607 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 16 |
Hb_004242_120 |
0.250581858 |
- |
- |
Organic cation transporter, putative [Ricinus communis] |
| 17 |
Hb_007317_130 |
0.2508039852 |
- |
- |
retrotransposon protein, putative, unclassified [Oryza sativa Japonica Group] |
| 18 |
Hb_018105_010 |
0.2514782745 |
- |
- |
hypothetical protein POPTR_0004s08710g [Populus trichocarpa] |
| 19 |
Hb_042463_050 |
0.2543966954 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_002030_050 |
0.2546943523 |
- |
- |
classical arabinogalactan protein 7 precursor [Jatropha curcas] |