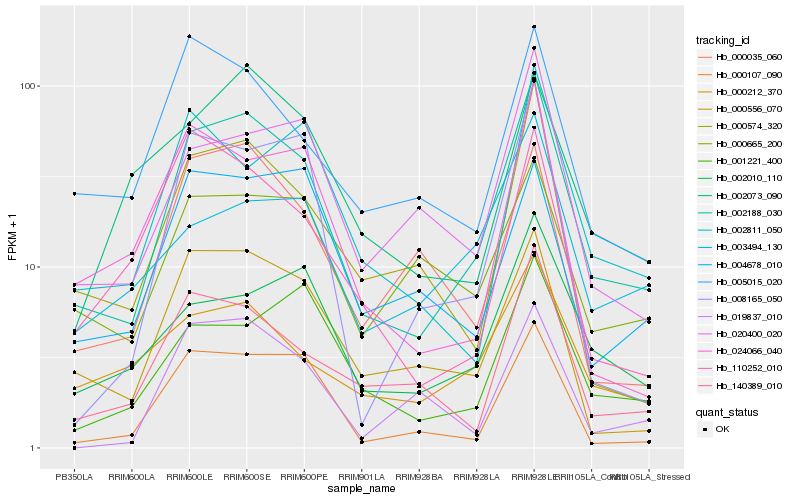
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002188_030 |
0.0 |
- |
- |
PREDICTED: probable zinc metallopeptidase EGY3, chloroplastic isoform X1 [Jatropha curcas] |
| 2 |
Hb_000556_070 |
0.1568926328 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase At2g45590 [Jatropha curcas] |
| 3 |
Hb_000107_090 |
0.1589231093 |
- |
- |
PREDICTED: kinesin KP1 [Jatropha curcas] |
| 4 |
Hb_000574_320 |
0.165437826 |
- |
- |
hypothetical protein CICLE_v10024937mg [Citrus clementina] |
| 5 |
Hb_140389_010 |
0.1666159064 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 6 |
Hb_020400_020 |
0.1723761237 |
- |
- |
starch synthase isoform II [Manihot esculenta] |
| 7 |
Hb_005015_020 |
0.1749819963 |
desease resistance |
Gene Name: ABC_tran |
ABC transporter family protein [Hevea brasiliensis] |
| 8 |
Hb_002010_110 |
0.1757431743 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1 [Jatropha curcas] |
| 9 |
Hb_024066_040 |
0.1763659609 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 10 |
Hb_000212_370 |
0.1808367783 |
- |
- |
thioredoxin f-type, putative [Ricinus communis] |
| 11 |
Hb_001221_400 |
0.1834308906 |
- |
- |
PREDICTED: CBS domain-containing protein CBSCBSPB1 [Jatropha curcas] |
| 12 |
Hb_008165_050 |
0.183523446 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 13 |
Hb_000665_200 |
0.1836361613 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 14 |
Hb_002811_050 |
0.183666311 |
- |
- |
PREDICTED: uncharacterized protein LOC105647652 [Jatropha curcas] |
| 15 |
Hb_000035_060 |
0.1842696037 |
- |
- |
PREDICTED: chaperone protein ClpB4, mitochondrial [Jatropha curcas] |
| 16 |
Hb_002073_090 |
0.1871643273 |
- |
- |
PREDICTED: copper transporter 6-like [Jatropha curcas] |
| 17 |
Hb_004678_010 |
0.187774199 |
transcription factor |
TF Family: Orphans |
Salt-tolerance protein, putative [Ricinus communis] |
| 18 |
Hb_110252_010 |
0.1879914731 |
- |
- |
hypothetical protein JCGZ_16705 [Jatropha curcas] |
| 19 |
Hb_019837_010 |
0.1891991946 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_003494_130 |
0.1896351446 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 9-like [Jatropha curcas] |