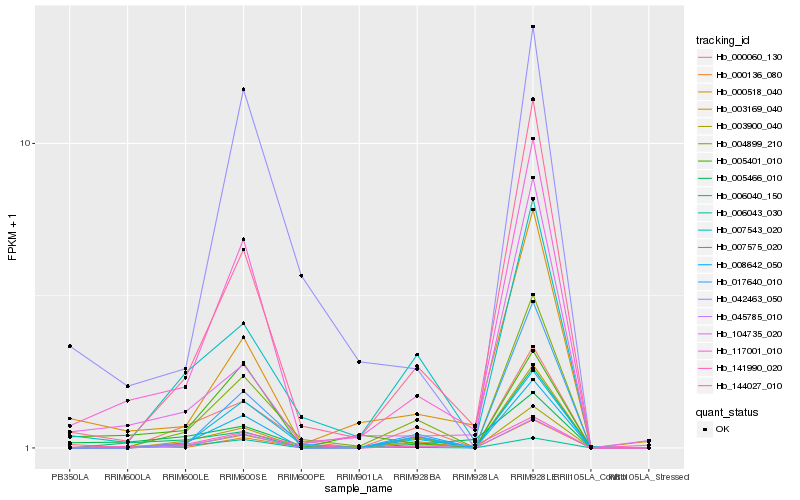
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_117001_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_144027_010 |
0.1299723353 |
- |
- |
- |
| 3 |
Hb_000518_040 |
0.154435408 |
- |
- |
- |
| 4 |
Hb_045785_010 |
0.174635786 |
- |
- |
PREDICTED: uncharacterized protein LOC105631130 [Jatropha curcas] |
| 5 |
Hb_004899_210 |
0.1780205729 |
- |
- |
PREDICTED: respiratory burst oxidase homolog protein E [Jatropha curcas] |
| 6 |
Hb_007575_020 |
0.1975564107 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 7 |
Hb_104735_020 |
0.1981460782 |
- |
- |
ATPase subunit 4 (mitochondrion) [Hevea brasiliensis] |
| 8 |
Hb_003900_040 |
0.1986725165 |
- |
- |
hypothetical protein [Lotus japonicus] |
| 9 |
Hb_008642_050 |
0.2055698138 |
- |
- |
- |
| 10 |
Hb_017640_010 |
0.2079834968 |
- |
- |
hypothetical protein JCGZ_18815 [Jatropha curcas] |
| 11 |
Hb_006043_030 |
0.2098663189 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 12 |
Hb_042463_050 |
0.2108633008 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_007543_020 |
0.2121033555 |
- |
- |
PREDICTED: uncharacterized protein LOC104229533, partial [Nicotiana sylvestris] |
| 14 |
Hb_006040_150 |
0.2123413057 |
- |
- |
- |
| 15 |
Hb_005401_010 |
0.2137239078 |
- |
- |
- |
| 16 |
Hb_005466_010 |
0.2154336341 |
- |
- |
ATPase subunit 6, partial (mitochondrion) [Celastrus scandens] |
| 17 |
Hb_000136_080 |
0.2170716323 |
- |
- |
hypothetical protein AALP_AA8G499800 [Arabis alpina] |
| 18 |
Hb_000060_130 |
0.2175453883 |
- |
- |
- |
| 19 |
Hb_141990_020 |
0.2201478232 |
- |
- |
RecName: Full=Genome polyprotein; Contains: RecName: Full=Movement protein; Short=MP; Contains: RecName: Full=Capsid protein; Short=CP; Contains: RecName: Full=Aspartic protease; Short=PR; Contains: RecName: Full=Reverse transcriptase; Short=RT [Petunia vein clearing virus isolate Hohn] |
| 20 |
Hb_003169_040 |
0.2235691854 |
- |
- |
- |