| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
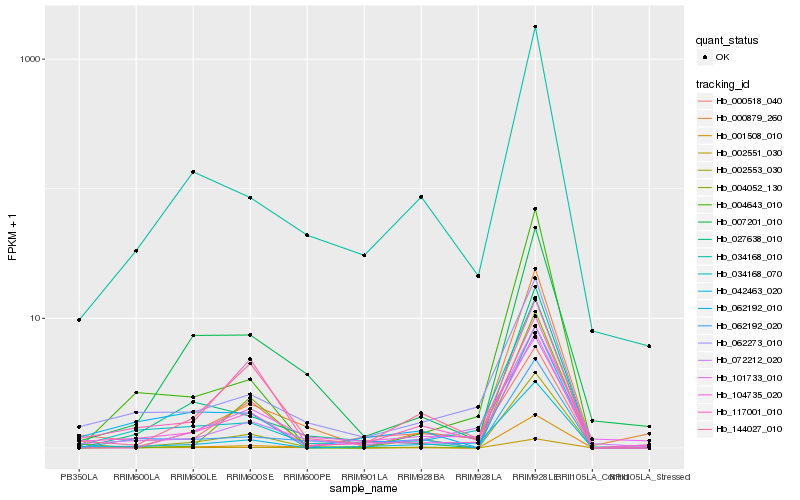
Hb_104735_020 |
0.0 |
- |
- |
ATPase subunit 4 (mitochondrion) [Hevea brasiliensis] |
| 2 |
Hb_042463_020 |
0.1363999128 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_034168_070 |
0.1444983352 |
- |
- |
ATP synthase F1 subunit 1 (mitochondrion) [Hevea brasiliensis] |
| 4 |
Hb_062273_010 |
0.146712489 |
- |
- |
ATP synthase F0 subunit 9 [Chara vulgaris] |
| 5 |
Hb_000518_040 |
0.1521655548 |
- |
- |
- |
| 6 |
Hb_072212_020 |
0.1575565806 |
- |
- |
hypothetical protein B456_001G162300 [Gossypium raimondii] |
| 7 |
Hb_034168_010 |
0.169983875 |
- |
- |
hypothetical protein [Azospirillum sp. CAG:260] |
| 8 |
Hb_002553_030 |
0.1709919783 |
- |
- |
hypothetical protein POPTR_0008s16550g [Populus trichocarpa] |
| 9 |
Hb_062192_010 |
0.1738814085 |
- |
- |
- |
| 10 |
Hb_144027_010 |
0.1790199755 |
- |
- |
- |
| 11 |
Hb_002551_030 |
0.1813562157 |
- |
- |
gypsy/Ty-3 retroelement polyprotein; 69905-74404 [Arabidopsis thaliana] |
| 12 |
Hb_101733_010 |
0.1867148159 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_027638_010 |
0.1890230827 |
- |
- |
Protein KES1, putative [Ricinus communis] |
| 14 |
Hb_007201_010 |
0.1970161316 |
- |
- |
PREDICTED: uncharacterized protein LOC105631226 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000879_260 |
0.1977532153 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At5g43190 [Jatropha curcas] |
| 16 |
Hb_117001_010 |
0.1981460782 |
- |
- |
- |
| 17 |
Hb_004052_130 |
0.1982336148 |
- |
- |
PREDICTED: cytochrome P450 78A5 [Jatropha curcas] |
| 18 |
Hb_004643_010 |
0.1986806019 |
- |
- |
cytochrome B, putative [Ricinus communis] |
| 19 |
Hb_062192_020 |
0.1987467305 |
- |
- |
cytochrome c biogenesis FN (mitochondrion) [Hevea brasiliensis] |
| 20 |
Hb_001508_010 |
0.2002729888 |
- |
- |
conserved hypothetical protein [Ricinus communis] |