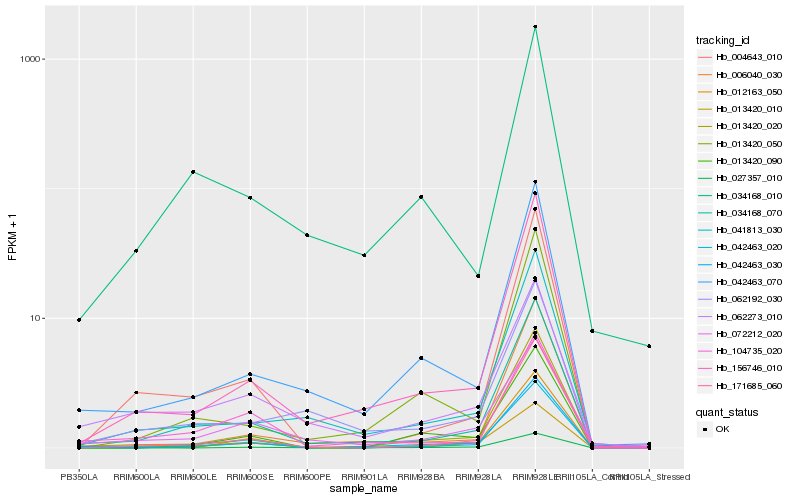
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_042463_020 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_034168_070 |
0.1117766396 |
- |
- |
ATP synthase F1 subunit 1 (mitochondrion) [Hevea brasiliensis] |
| 3 |
Hb_156746_010 |
0.133426664 |
- |
- |
ATP synthase F1 subunit 1 (mitochondrion) [Hevea brasiliensis] |
| 4 |
Hb_012163_050 |
0.1338111411 |
- |
- |
ATPase subunit [Medicago truncatula] |
| 5 |
Hb_104735_020 |
0.1363999128 |
- |
- |
ATPase subunit 4 (mitochondrion) [Hevea brasiliensis] |
| 6 |
Hb_062273_010 |
0.1379481471 |
- |
- |
ATP synthase F0 subunit 9 [Chara vulgaris] |
| 7 |
Hb_042463_070 |
0.139689273 |
- |
- |
60S ribosomal protein L16, putative [Ricinus communis] |
| 8 |
Hb_072212_020 |
0.1420614337 |
- |
- |
hypothetical protein B456_001G162300 [Gossypium raimondii] |
| 9 |
Hb_013420_010 |
0.1470860887 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_013420_020 |
0.1505641744 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_013420_090 |
0.1517893837 |
- |
- |
BnaCnng13090D [Brassica napus] |
| 12 |
Hb_004643_010 |
0.1613171389 |
- |
- |
cytochrome B, putative [Ricinus communis] |
| 13 |
Hb_041813_030 |
0.1645901104 |
- |
- |
cytochrome oxidase subunit 1, partial (mitochondrion) [Calochone redingii] |
| 14 |
Hb_042463_030 |
0.1646195976 |
- |
- |
unknown [Lotus japonicus] |
| 15 |
Hb_171685_060 |
0.1652823635 |
- |
- |
PREDICTED: cytochrome c oxidase subunit 2 [Pyrus x bretschneideri] |
| 16 |
Hb_034168_010 |
0.1691582859 |
- |
- |
hypothetical protein [Azospirillum sp. CAG:260] |
| 17 |
Hb_006040_030 |
0.1698083582 |
- |
- |
hypothetical protein EUTSA_v10001274mg [Eutrema salsugineum] |
| 18 |
Hb_013420_050 |
0.1728677839 |
- |
- |
hypothetical protein (mitochondrion) [Vicia faba] |
| 19 |
Hb_062192_030 |
0.1739233031 |
- |
- |
ribosomal protein S1 (mitochondrion) [Hevea brasiliensis] |
| 20 |
Hb_027357_010 |
0.1759378481 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |