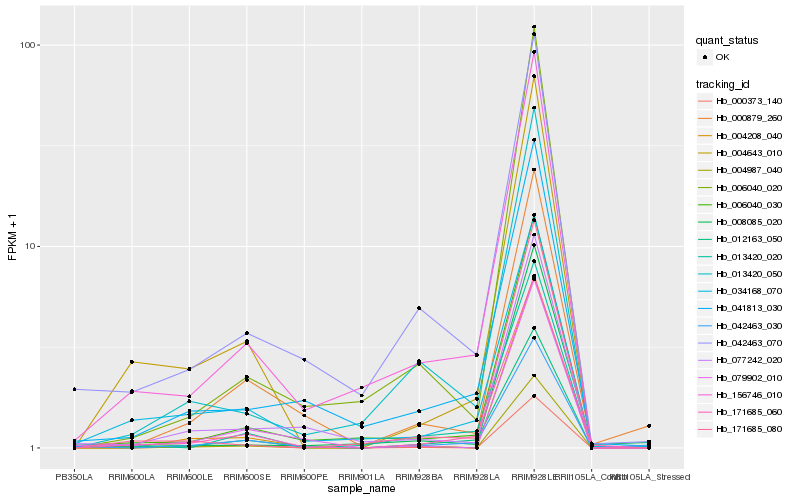
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_171685_060 |
0.0 |
- |
- |
PREDICTED: cytochrome c oxidase subunit 2 [Pyrus x bretschneideri] |
| 2 |
Hb_012163_050 |
0.0787985037 |
- |
- |
ATPase subunit [Medicago truncatula] |
| 3 |
Hb_156746_010 |
0.1036812705 |
- |
- |
ATP synthase F1 subunit 1 (mitochondrion) [Hevea brasiliensis] |
| 4 |
Hb_004643_010 |
0.1050882604 |
- |
- |
cytochrome B, putative [Ricinus communis] |
| 5 |
Hb_006040_030 |
0.1068981037 |
- |
- |
hypothetical protein EUTSA_v10001274mg [Eutrema salsugineum] |
| 6 |
Hb_013420_020 |
0.1164328391 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_041813_030 |
0.123797679 |
- |
- |
cytochrome oxidase subunit 1, partial (mitochondrion) [Calochone redingii] |
| 8 |
Hb_042463_030 |
0.1251607159 |
- |
- |
unknown [Lotus japonicus] |
| 9 |
Hb_004208_040 |
0.1263874819 |
- |
- |
UDP-glycosyltransferase 86A1 [Morus notabilis] |
| 10 |
Hb_171685_080 |
0.1272520915 |
- |
- |
unknown [Allium cepa] |
| 11 |
Hb_006040_020 |
0.1320302727 |
- |
- |
ATPase subunit 8 (mitochondrion) [Hevea brasiliensis] |
| 12 |
Hb_013420_050 |
0.1330496664 |
- |
- |
hypothetical protein (mitochondrion) [Vicia faba] |
| 13 |
Hb_034168_070 |
0.1344264408 |
- |
- |
ATP synthase F1 subunit 1 (mitochondrion) [Hevea brasiliensis] |
| 14 |
Hb_000373_140 |
0.1381425252 |
- |
- |
PREDICTED: uncharacterized protein LOC105160092 [Sesamum indicum] |
| 15 |
Hb_004987_040 |
0.1383725143 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 16 |
Hb_000879_260 |
0.141475738 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At5g43190 [Jatropha curcas] |
| 17 |
Hb_008085_020 |
0.1422718359 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g49770 [Populus euphratica] |
| 18 |
Hb_042463_070 |
0.143376638 |
- |
- |
60S ribosomal protein L16, putative [Ricinus communis] |
| 19 |
Hb_079902_010 |
0.1478258698 |
- |
- |
cytochrome c biogenesis B (mitochondrion) [Hevea brasiliensis] |
| 20 |
Hb_077242_020 |
0.1483727926 |
- |
- |
ribosomal protein S4 (mitochondrion) [Ricinus communis] |