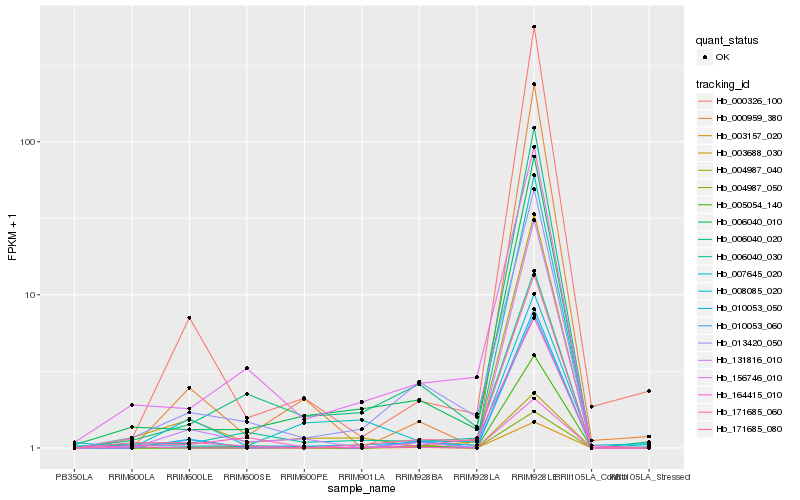
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_171685_080 |
0.0 |
- |
- |
unknown [Allium cepa] |
| 2 |
Hb_006040_010 |
0.08312612 |
- |
- |
cytochrome c oxidase subunit 3 (mitochondrion) [Hevea brasiliensis] |
| 3 |
Hb_006040_020 |
0.0865545398 |
- |
- |
ATPase subunit 8 (mitochondrion) [Hevea brasiliensis] |
| 4 |
Hb_003688_030 |
0.0938650928 |
- |
- |
NADH dehydrogenase 2-like ORF 180 |
| 5 |
Hb_013420_050 |
0.0973351332 |
- |
- |
hypothetical protein (mitochondrion) [Vicia faba] |
| 6 |
Hb_008085_020 |
0.0975077788 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g49770 [Populus euphratica] |
| 7 |
Hb_006040_030 |
0.1054490997 |
- |
- |
hypothetical protein EUTSA_v10001274mg [Eutrema salsugineum] |
| 8 |
Hb_003157_020 |
0.1061911418 |
- |
- |
PREDICTED: uncharacterized protein LOC104809111 [Tarenaya hassleriana] |
| 9 |
Hb_164415_010 |
0.1063070533 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000326_100 |
0.1105047566 |
transcription factor |
TF Family: MYB-related |
DnaJ homolog subfamily C member 2 [Morus notabilis] |
| 11 |
Hb_156746_010 |
0.1167603496 |
- |
- |
ATP synthase F1 subunit 1 (mitochondrion) [Hevea brasiliensis] |
| 12 |
Hb_007645_020 |
0.1179304749 |
- |
- |
NAD(P)H-quinone oxidoreductase subunit [Medicago truncatula] |
| 13 |
Hb_010053_060 |
0.1182023939 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 14 |
Hb_131816_010 |
0.1186295553 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 15 |
Hb_010053_050 |
0.1188430874 |
- |
- |
hypothetical protein POPTR_0530s002102g, partial [Populus trichocarpa] |
| 16 |
Hb_004987_050 |
0.1194570653 |
- |
- |
- |
| 17 |
Hb_004987_040 |
0.1199440164 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 18 |
Hb_000959_380 |
0.1227144721 |
- |
- |
PREDICTED: (-)-alpha-terpineol synthase-like [Vitis vinifera] |
| 19 |
Hb_005054_140 |
0.1251180334 |
- |
- |
PREDICTED: laccase-4-like [Jatropha curcas] |
| 20 |
Hb_171685_060 |
0.1272520915 |
- |
- |
PREDICTED: cytochrome c oxidase subunit 2 [Pyrus x bretschneideri] |