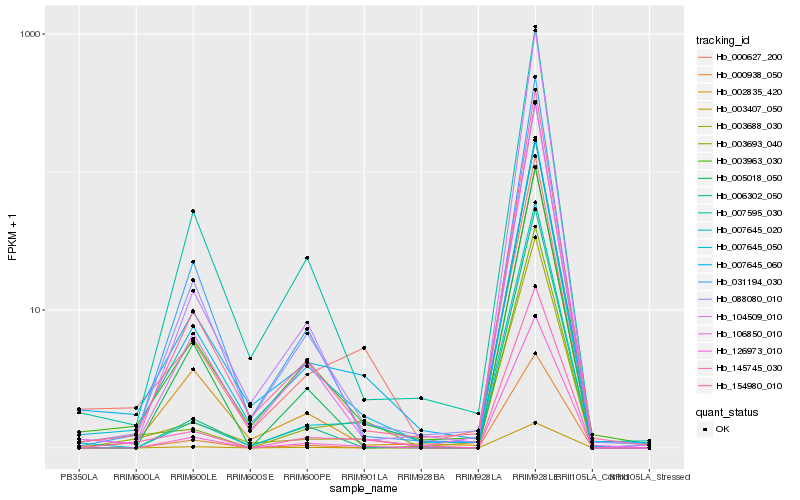
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_145745_030 |
0.0 |
- |
- |
BnaC09g26890D [Brassica napus] |
| 2 |
Hb_003693_040 |
0.0706330626 |
- |
- |
NADH dehydrogenase subunit 1 [Hevea brasiliensis] |
| 3 |
Hb_003688_030 |
0.0768016663 |
- |
- |
NADH dehydrogenase 2-like ORF 180 |
| 4 |
Hb_126973_010 |
0.080529738 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g34110 [Vitis vinifera] |
| 5 |
Hb_007645_050 |
0.0811453585 |
- |
- |
acetyl-CoA carboxylase beta subunit [Hevea brasiliensis] |
| 6 |
Hb_003963_030 |
0.083305445 |
- |
- |
psbB [Jatropha curcas] |
| 7 |
Hb_002835_420 |
0.0910199602 |
- |
- |
photosystem I P700 chlorophyll A apoprotein A2 [Medicago truncatula] |
| 8 |
Hb_154980_010 |
0.0913605838 |
- |
- |
photosystem I P700 chlorophyll A apoprotein A2 [Medicago truncatula] |
| 9 |
Hb_007645_020 |
0.0921510009 |
- |
- |
NAD(P)H-quinone oxidoreductase subunit [Medicago truncatula] |
| 10 |
Hb_007645_060 |
0.0974478151 |
- |
- |
cytochrome f [Hevea brasiliensis] |
| 11 |
Hb_106850_010 |
0.0977883332 |
- |
- |
photosystem II protein D2 [Lactuca sativa] |
| 12 |
Hb_088080_010 |
0.0999929815 |
- |
- |
photosystem I P700 chlorophyll a apoprotein A1 [Matelea biflora] |
| 13 |
Hb_031194_030 |
0.1016650649 |
- |
- |
ribulose 1,5-bisphosphate carboxylase/oxygenase [Stackhousia minima] |
| 14 |
Hb_000938_050 |
0.1031868339 |
- |
- |
hypothetical protein JCGZ_09640 [Jatropha curcas] |
| 15 |
Hb_007595_030 |
0.1048321149 |
- |
- |
psbA gene product (chloroplast) [Silene noctiflora] |
| 16 |
Hb_000627_200 |
0.1054180595 |
- |
- |
NADH dehydrogenase subunit I [Hevea brasiliensis] |
| 17 |
Hb_006302_050 |
0.1074258406 |
- |
- |
PREDICTED: uncharacterized protein LOC104804969, partial [Tarenaya hassleriana] |
| 18 |
Hb_104509_010 |
0.107598405 |
- |
- |
photosystem II CP43 chlorophyll apoprotein (chloroplast) [Ficus sp. Moore 315] |
| 19 |
Hb_003407_050 |
0.1085661445 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 20 |
Hb_005018_050 |
0.1089794699 |
- |
- |
PREDICTED: licodione synthase-like [Jatropha curcas] |