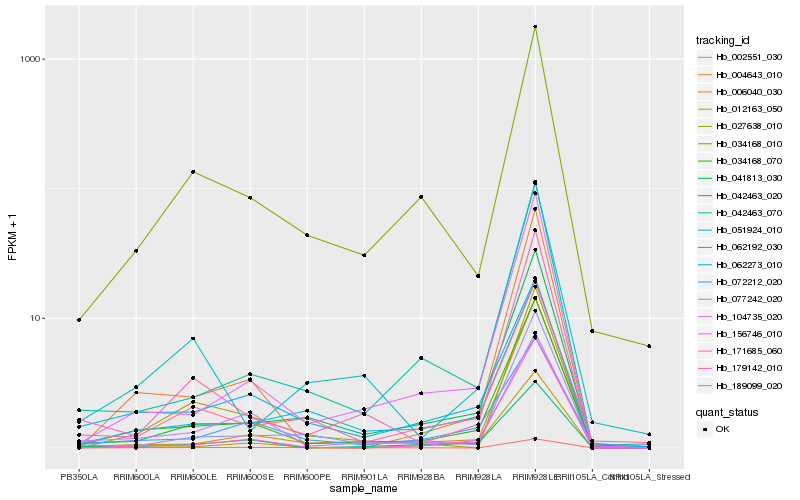
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_034168_070 |
0.0 |
- |
- |
ATP synthase F1 subunit 1 (mitochondrion) [Hevea brasiliensis] |
| 2 |
Hb_156746_010 |
0.1088479125 |
- |
- |
ATP synthase F1 subunit 1 (mitochondrion) [Hevea brasiliensis] |
| 3 |
Hb_042463_020 |
0.1117766396 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_004643_010 |
0.1136396227 |
- |
- |
cytochrome B, putative [Ricinus communis] |
| 5 |
Hb_041813_030 |
0.1258973141 |
- |
- |
cytochrome oxidase subunit 1, partial (mitochondrion) [Calochone redingii] |
| 6 |
Hb_042463_070 |
0.127795004 |
- |
- |
60S ribosomal protein L16, putative [Ricinus communis] |
| 7 |
Hb_012163_050 |
0.1316890238 |
- |
- |
ATPase subunit [Medicago truncatula] |
| 8 |
Hb_062192_030 |
0.1326897101 |
- |
- |
ribosomal protein S1 (mitochondrion) [Hevea brasiliensis] |
| 9 |
Hb_171685_060 |
0.1344264408 |
- |
- |
PREDICTED: cytochrome c oxidase subunit 2 [Pyrus x bretschneideri] |
| 10 |
Hb_006040_030 |
0.1364040507 |
- |
- |
hypothetical protein EUTSA_v10001274mg [Eutrema salsugineum] |
| 11 |
Hb_062273_010 |
0.1380661448 |
- |
- |
ATP synthase F0 subunit 9 [Chara vulgaris] |
| 12 |
Hb_179142_010 |
0.1388184602 |
- |
- |
NADH dehydrogenase ND 2 subunit [Nicotiana tomentosiformis] |
| 13 |
Hb_189099_020 |
0.1394638121 |
- |
- |
NADH dehydrogenase subunit 9 (mitochondrion) [Hevea brasiliensis] |
| 14 |
Hb_034168_010 |
0.140061232 |
- |
- |
hypothetical protein [Azospirillum sp. CAG:260] |
| 15 |
Hb_077242_020 |
0.1413613377 |
- |
- |
ribosomal protein S4 (mitochondrion) [Ricinus communis] |
| 16 |
Hb_104735_020 |
0.1444983352 |
- |
- |
ATPase subunit 4 (mitochondrion) [Hevea brasiliensis] |
| 17 |
Hb_072212_020 |
0.1457916734 |
- |
- |
hypothetical protein B456_001G162300 [Gossypium raimondii] |
| 18 |
Hb_027638_010 |
0.1483381105 |
- |
- |
Protein KES1, putative [Ricinus communis] |
| 19 |
Hb_051924_010 |
0.1529159492 |
- |
- |
BnaA01g34100D [Brassica napus] |
| 20 |
Hb_002551_030 |
0.1537838013 |
- |
- |
gypsy/Ty-3 retroelement polyprotein; 69905-74404 [Arabidopsis thaliana] |