| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
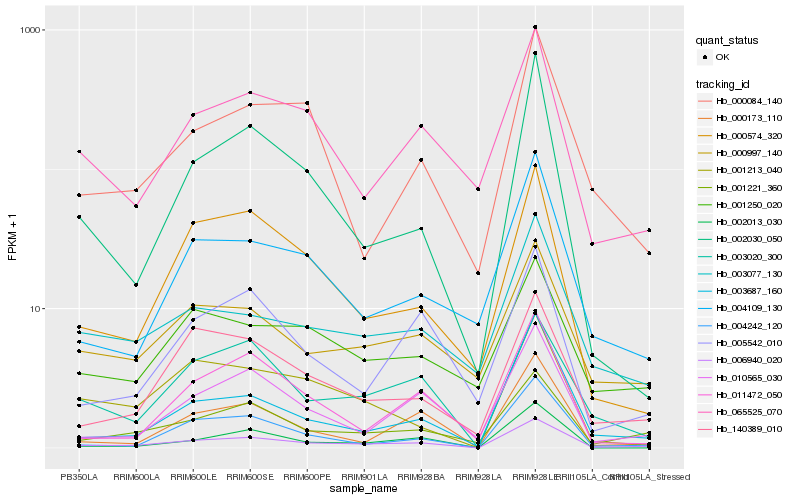
Hb_006940_020 |
0.0 |
- |
- |
polyprotein [Oryza australiensis] |
| 2 |
Hb_001221_360 |
0.1267796226 |
- |
- |
late embryogenesis abundant, putative [Ricinus communis] |
| 3 |
Hb_010565_030 |
0.1497178366 |
- |
- |
hypothetical protein VITISV_006810 [Vitis vinifera] |
| 4 |
Hb_003687_160 |
0.1522882402 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 5 |
Hb_003020_300 |
0.1555591 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000173_110 |
0.1568898174 |
- |
- |
putative RNA-directed DNA polymerase (Reverse transcriptase) [Malus domestica] |
| 7 |
Hb_004242_120 |
0.1591509132 |
- |
- |
Organic cation transporter, putative [Ricinus communis] |
| 8 |
Hb_000997_140 |
0.1613192538 |
- |
- |
PREDICTED: chloroplastic group IIA intron splicing facilitator CRS1, chloroplastic [Jatropha curcas] |
| 9 |
Hb_140389_010 |
0.1616005577 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 10 |
Hb_004109_130 |
0.1630740198 |
- |
- |
PREDICTED: translation initiation factor IF-2, chloroplastic [Jatropha curcas] |
| 11 |
Hb_065525_070 |
0.163536552 |
- |
- |
unnamed protein product [Homo sapiens] |
| 12 |
Hb_002030_050 |
0.1643692475 |
- |
- |
classical arabinogalactan protein 7 precursor [Jatropha curcas] |
| 13 |
Hb_003077_130 |
0.1655585226 |
- |
- |
hypothetical protein JCGZ_22476 [Jatropha curcas] |
| 14 |
Hb_000574_320 |
0.1667062112 |
- |
- |
hypothetical protein CICLE_v10024937mg [Citrus clementina] |
| 15 |
Hb_002013_030 |
0.1695503508 |
- |
- |
PREDICTED: uncharacterized protein LOC104799692 [Tarenaya hassleriana] |
| 16 |
Hb_011472_050 |
0.1722589857 |
- |
- |
- |
| 17 |
Hb_005542_010 |
0.1723992246 |
- |
- |
Uncharacterized protein TCM_021447 [Theobroma cacao] |
| 18 |
Hb_000084_140 |
0.1767885185 |
- |
- |
Aspartic proteinase nepenthesin-2 precursor, putative [Ricinus communis] |
| 19 |
Hb_001213_040 |
0.1773486311 |
- |
- |
fructose-1,6-bisphosphatase, putative [Ricinus communis] |
| 20 |
Hb_001250_020 |
0.1776478001 |
- |
- |
PREDICTED: CRS2-associated factor 2, chloroplastic [Jatropha curcas] |