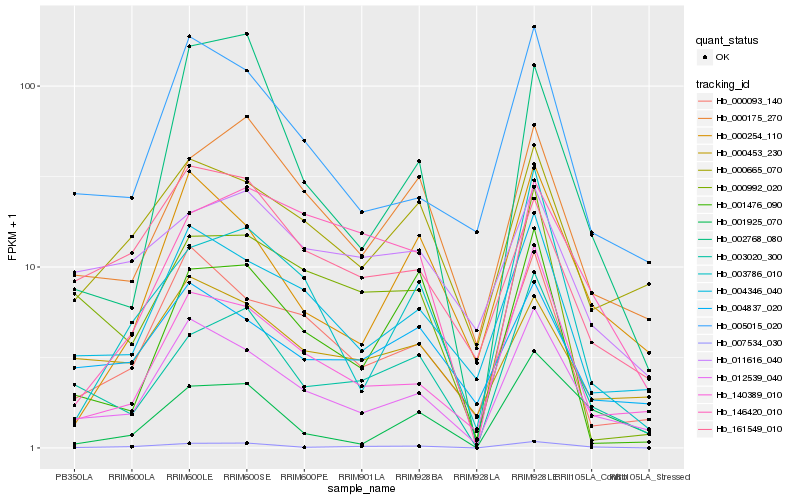
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007534_030 |
0.0 |
- |
- |
PREDICTED: receptor-like protein kinase [Jatropha curcas] |
| 2 |
Hb_003020_300 |
0.164289206 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_012539_040 |
0.1730462974 |
transcription factor |
TF Family: CPP |
hypothetical protein JCGZ_07765 [Jatropha curcas] |
| 4 |
Hb_000175_270 |
0.2144086909 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 11 isoform X1 [Jatropha curcas] |
| 5 |
Hb_140389_010 |
0.217177016 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 6 |
Hb_002768_080 |
0.217643727 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranyl-diphosphate synthase [Hevea brasiliensis] |
| 7 |
Hb_161549_010 |
0.2206494534 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000254_110 |
0.2209290833 |
- |
- |
PREDICTED: dynein light chain 1, cytoplasmic-like [Jatropha curcas] |
| 9 |
Hb_000665_070 |
0.2222899449 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein knotted-1-like 3 isoform X1 [Jatropha curcas] |
| 10 |
Hb_005015_020 |
0.2230936049 |
desease resistance |
Gene Name: ABC_tran |
ABC transporter family protein [Hevea brasiliensis] |
| 11 |
Hb_004837_020 |
0.2248815273 |
- |
- |
PREDICTED: uncharacterized protein LOC105648271 [Jatropha curcas] |
| 12 |
Hb_004346_040 |
0.2260599335 |
- |
- |
hypothetical protein POPTR_0016s06630g [Populus trichocarpa] |
| 13 |
Hb_001925_070 |
0.2269282718 |
- |
- |
PREDICTED: 40S ribosomal protein S3-3-like [Populus euphratica] |
| 14 |
Hb_003786_010 |
0.2272814659 |
transcription factor |
TF Family: HB |
PREDICTED: BEL1-like homeodomain protein 2 isoform X2 [Jatropha curcas] |
| 15 |
Hb_011616_040 |
0.228225152 |
- |
- |
glucan water dikinase 3, partial [Manihot esculenta] |
| 16 |
Hb_146420_010 |
0.2284285756 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 17 |
Hb_000093_140 |
0.2319896511 |
- |
- |
PREDICTED: fructokinase-like 2, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000453_230 |
0.233479151 |
- |
- |
PREDICTED: NO-associated protein 1, chloroplastic/mitochondrial [Jatropha curcas] |
| 19 |
Hb_000992_020 |
0.2354315504 |
- |
- |
monoxygenase, putative [Ricinus communis] |
| 20 |
Hb_001476_090 |
0.2358295334 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 3 [Jatropha curcas] |