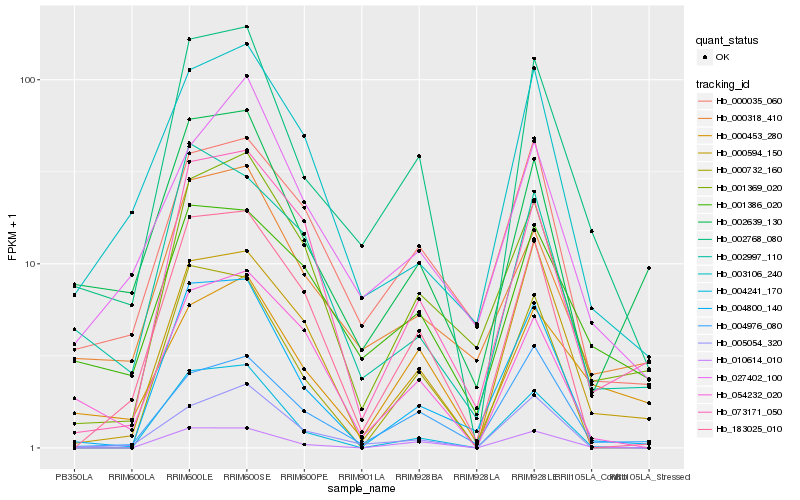
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002768_080 |
0.0 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranyl-diphosphate synthase [Hevea brasiliensis] |
| 2 |
Hb_010614_010 |
0.1601641514 |
- |
- |
leucine-rich repeat transmembrane protein kinase [Populus trichocarpa] |
| 3 |
Hb_001386_020 |
0.1655969693 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_004800_140 |
0.1672087634 |
- |
- |
PREDICTED: uncharacterized protein LOC105633940 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000318_410 |
0.1682611795 |
- |
- |
PREDICTED: extra-large guanine nucleotide-binding protein 1-like [Jatropha curcas] |
| 6 |
Hb_005054_320 |
0.1686989576 |
- |
- |
PREDICTED: uncharacterized protein LOC105646518 [Jatropha curcas] |
| 7 |
Hb_000594_150 |
0.1698652089 |
- |
- |
PREDICTED: serine/threonine-protein kinase STN7, chloroplastic-like isoform X1 [Nelumbo nucifera] |
| 8 |
Hb_000453_280 |
0.1703679203 |
- |
- |
negative cofactor 2 transcriptional co-repressor, putative [Ricinus communis] |
| 9 |
Hb_004976_080 |
0.1714798543 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647845 [Jatropha curcas] |
| 10 |
Hb_000035_060 |
0.1737988507 |
- |
- |
PREDICTED: chaperone protein ClpB4, mitochondrial [Jatropha curcas] |
| 11 |
Hb_001369_020 |
0.1759165524 |
- |
- |
PREDICTED: U-box domain-containing protein 35-like [Jatropha curcas] |
| 12 |
Hb_183025_010 |
0.1760739398 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA1H1 [Jatropha curcas] |
| 13 |
Hb_003106_240 |
0.1789299433 |
- |
- |
hypothetical protein POPTR_0012s07880g [Populus trichocarpa] |
| 14 |
Hb_054232_020 |
0.1798680039 |
- |
- |
Potassium channel SKOR, putative [Ricinus communis] |
| 15 |
Hb_027402_100 |
0.1812877322 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein JCGZ_11638 [Jatropha curcas] |
| 16 |
Hb_002639_130 |
0.1858917443 |
- |
- |
hypothetical protein JCGZ_08129 [Jatropha curcas] |
| 17 |
Hb_002997_110 |
0.1866635849 |
transcription factor |
TF Family: Orphans |
PREDICTED: B-box zinc finger protein 22 isoform X1 [Jatropha curcas] |
| 18 |
Hb_073171_050 |
0.1870156085 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-like protein H [Jatropha curcas] |
| 19 |
Hb_004241_170 |
0.1881277073 |
- |
- |
PREDICTED: uncharacterized protein LOC105633220 [Jatropha curcas] |
| 20 |
Hb_000732_160 |
0.1915926576 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB3-like [Jatropha curcas] |