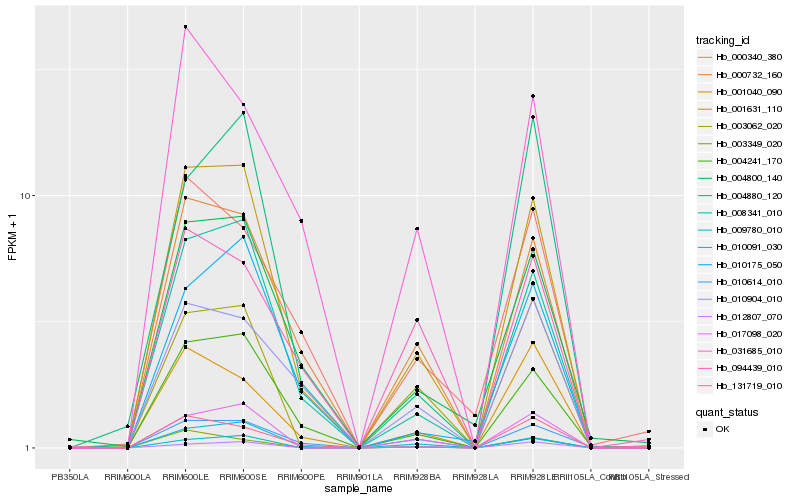
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001631_110 |
0.0 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: gamma-glutamyltranspeptidase 1-like [Populus euphratica] |
| 2 |
Hb_008341_010 |
0.0747593282 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein HD1-like isoform X2 [Jatropha curcas] |
| 3 |
Hb_004241_170 |
0.0895894869 |
- |
- |
PREDICTED: uncharacterized protein LOC105633220 [Jatropha curcas] |
| 4 |
Hb_000732_160 |
0.1030086872 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB3-like [Jatropha curcas] |
| 5 |
Hb_010091_030 |
0.1180446079 |
- |
- |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 6 |
Hb_004800_140 |
0.1272567255 |
- |
- |
PREDICTED: uncharacterized protein LOC105633940 isoform X1 [Jatropha curcas] |
| 7 |
Hb_017098_020 |
0.1348048921 |
- |
- |
Zinc finger protein GIS2 [Medicago truncatula] |
| 8 |
Hb_010614_010 |
0.1356358433 |
- |
- |
leucine-rich repeat transmembrane protein kinase [Populus trichocarpa] |
| 9 |
Hb_010175_050 |
0.1513059979 |
- |
- |
PREDICTED: uncharacterized protein LOC105639906 [Jatropha curcas] |
| 10 |
Hb_003349_020 |
0.1525338621 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein At4g27190-like [Beta vulgaris subsp. vulgaris] |
| 11 |
Hb_001040_090 |
0.1546791715 |
- |
- |
PREDICTED: tetraspanin-19-like [Jatropha curcas] |
| 12 |
Hb_003062_020 |
0.1558397569 |
- |
- |
Polyprotein, putative [Solanum demissum] |
| 13 |
Hb_010904_010 |
0.1613743113 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g39030 isoform X2 [Jatropha curcas] |
| 14 |
Hb_031685_010 |
0.1688502282 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 15 |
Hb_000340_380 |
0.1689429276 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK2 [Jatropha curcas] |
| 16 |
Hb_131719_010 |
0.1725031553 |
- |
- |
PREDICTED: uncharacterized protein LOC104107451 [Nicotiana tomentosiformis] |
| 17 |
Hb_004880_120 |
0.172588905 |
- |
- |
PREDICTED: calcium-binding protein PBP1 [Jatropha curcas] |
| 18 |
Hb_094439_010 |
0.1730014814 |
- |
- |
PREDICTED: MACPF domain-containing protein At1g14780 [Jatropha curcas] |
| 19 |
Hb_012807_070 |
0.1731716507 |
- |
- |
PREDICTED: UDP-glycosyltransferase 86A2 [Jatropha curcas] |
| 20 |
Hb_009780_010 |
0.1750963108 |
- |
- |
conserved hypothetical protein [Ricinus communis] |