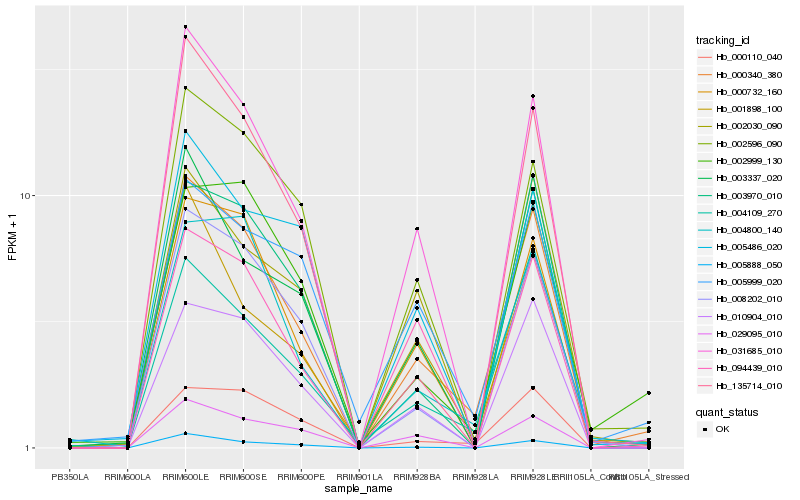
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000340_380 |
0.0 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK2 [Jatropha curcas] |
| 2 |
Hb_031685_010 |
0.1127069984 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 3 |
Hb_004109_270 |
0.1221433988 |
transcription factor |
TF Family: NAC |
transcription activator NAC1 family protein [Populus trichocarpa] |
| 4 |
Hb_010904_010 |
0.1256819392 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g39030 isoform X2 [Jatropha curcas] |
| 5 |
Hb_004800_140 |
0.127139587 |
- |
- |
PREDICTED: uncharacterized protein LOC105633940 isoform X1 [Jatropha curcas] |
| 6 |
Hb_008202_010 |
0.129104722 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 7 |
Hb_000732_160 |
0.1296934776 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB3-like [Jatropha curcas] |
| 8 |
Hb_000110_040 |
0.1314346063 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase CVP2 [Jatropha curcas] |
| 9 |
Hb_002596_090 |
0.1338678247 |
- |
- |
PREDICTED: calcium-transporting ATPase 4, plasma membrane-type-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_003337_020 |
0.1388087812 |
- |
- |
PREDICTED: uncharacterized protein LOC105116455 [Populus euphratica] |
| 11 |
Hb_003970_010 |
0.139872403 |
- |
- |
PREDICTED: sigma factor binding protein 1, chloroplastic-like [Populus euphratica] |
| 12 |
Hb_005888_050 |
0.1401027572 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 13 |
Hb_135714_010 |
0.1443515692 |
- |
- |
hypothetical protein POPTR_0017s09310g, partial [Populus trichocarpa] |
| 14 |
Hb_002999_130 |
0.1451421282 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein POPTR_0008s08740g [Populus trichocarpa] |
| 15 |
Hb_029095_010 |
0.1471432786 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_21060 [Jatropha curcas] |
| 16 |
Hb_094439_010 |
0.1497673104 |
- |
- |
PREDICTED: MACPF domain-containing protein At1g14780 [Jatropha curcas] |
| 17 |
Hb_001898_100 |
0.1510264869 |
- |
- |
PREDICTED: phosphatidylserine decarboxylase proenzyme 2-like [Jatropha curcas] |
| 18 |
Hb_005486_020 |
0.1524653544 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HAT5 [Jatropha curcas] |
| 19 |
Hb_002030_090 |
0.1536626393 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 20 |
Hb_005999_020 |
0.1536840032 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SENSITIVE TO PROTON RHIZOTOXICITY 1-like [Jatropha curcas] |