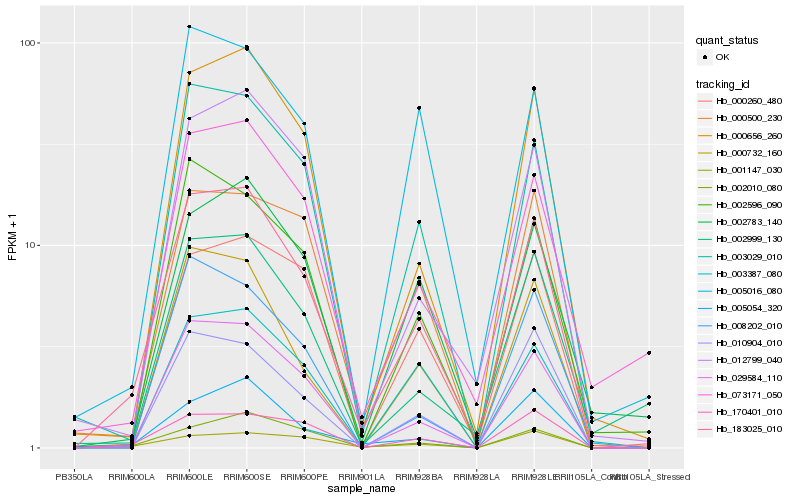
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_183025_010 |
0.0 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA1H1 [Jatropha curcas] |
| 2 |
Hb_029584_110 |
0.1020142031 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-like protein J [Jatropha curcas] |
| 3 |
Hb_003029_010 |
0.1166358453 |
transcription factor |
TF Family: WRKY |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000656_260 |
0.1190566407 |
- |
- |
PREDICTED: uncharacterized protein LOC105631376 [Jatropha curcas] |
| 5 |
Hb_170401_010 |
0.1328511474 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 6 |
Hb_000732_160 |
0.1346185488 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB3-like [Jatropha curcas] |
| 7 |
Hb_073171_050 |
0.1349191815 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-like protein H [Jatropha curcas] |
| 8 |
Hb_002596_090 |
0.13497297 |
- |
- |
PREDICTED: calcium-transporting ATPase 4, plasma membrane-type-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_002010_080 |
0.1350505538 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g11050 [Jatropha curcas] |
| 10 |
Hb_012799_040 |
0.1386066129 |
- |
- |
PREDICTED: transmembrane ascorbate ferrireductase 1 [Jatropha curcas] |
| 11 |
Hb_005054_320 |
0.1404310319 |
- |
- |
PREDICTED: uncharacterized protein LOC105646518 [Jatropha curcas] |
| 12 |
Hb_002783_140 |
0.1410034261 |
- |
- |
PREDICTED: uncharacterized protein LOC105635332 [Jatropha curcas] |
| 13 |
Hb_000260_480 |
0.1431333079 |
transcription factor |
TF Family: GRAS |
PREDICTED: protein SCARECROW [Jatropha curcas] |
| 14 |
Hb_000500_230 |
0.1432745 |
- |
- |
hypothetical protein RCOM_0634810 [Ricinus communis] |
| 15 |
Hb_003387_080 |
0.1440141481 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 16 |
Hb_002999_130 |
0.144728014 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein POPTR_0008s08740g [Populus trichocarpa] |
| 17 |
Hb_010904_010 |
0.1451387275 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g39030 isoform X2 [Jatropha curcas] |
| 18 |
Hb_005016_080 |
0.1471458508 |
- |
- |
PREDICTED: neurofilament heavy polypeptide-like [Jatropha curcas] |
| 19 |
Hb_008202_010 |
0.1492992659 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 20 |
Hb_001147_030 |
0.1506535303 |
- |
- |
hypothetical protein JCGZ_14809 [Jatropha curcas] |