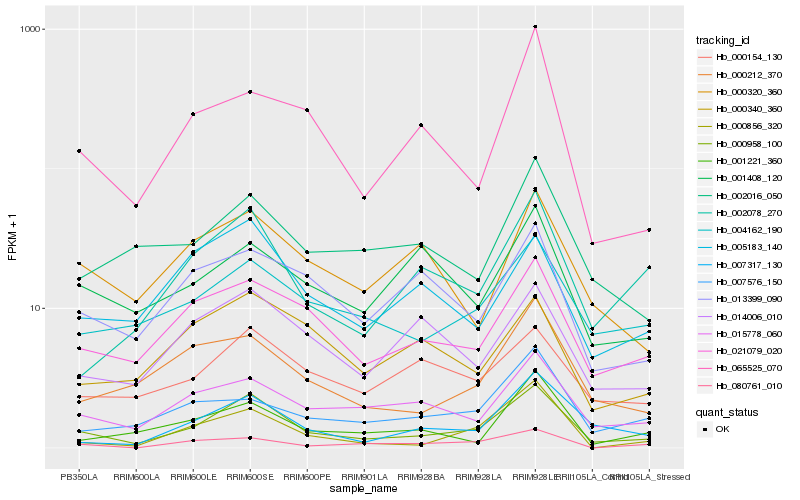
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000958_100 |
0.0 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 2 |
Hb_021079_020 |
0.1714640255 |
- |
- |
PREDICTED: uncharacterized protein LOC105648580 [Jatropha curcas] |
| 3 |
Hb_007317_130 |
0.1727523819 |
- |
- |
retrotransposon protein, putative, unclassified [Oryza sativa Japonica Group] |
| 4 |
Hb_015778_060 |
0.1802598444 |
- |
- |
PREDICTED: uncharacterized protein LOC105650696 [Jatropha curcas] |
| 5 |
Hb_000212_370 |
0.1822355571 |
- |
- |
thioredoxin f-type, putative [Ricinus communis] |
| 6 |
Hb_004162_190 |
0.1880698019 |
- |
- |
PREDICTED: uncharacterized protein LOC105627993 [Jatropha curcas] |
| 7 |
Hb_002078_270 |
0.1907844545 |
- |
- |
PREDICTED: F-box protein At1g67340 [Jatropha curcas] |
| 8 |
Hb_000856_320 |
0.1931571935 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin of 36 kDa-like [Jatropha curcas] |
| 9 |
Hb_065525_070 |
0.1968929504 |
- |
- |
unnamed protein product [Homo sapiens] |
| 10 |
Hb_005183_140 |
0.1988270849 |
- |
- |
PREDICTED: protein WEAK CHLOROPLAST MOVEMENT UNDER BLUE LIGHT 1-like [Jatropha curcas] |
| 11 |
Hb_014006_010 |
0.2005388166 |
- |
- |
PREDICTED: uncharacterized protein LOC105647962 [Jatropha curcas] |
| 12 |
Hb_000154_130 |
0.2014684783 |
- |
- |
PREDICTED: flowering time control protein FPA isoform X1 [Jatropha curcas] |
| 13 |
Hb_002016_050 |
0.2033593481 |
- |
- |
PREDICTED: translocase of chloroplast 159, chloroplastic [Jatropha curcas] |
| 14 |
Hb_001221_360 |
0.2052283338 |
- |
- |
late embryogenesis abundant, putative [Ricinus communis] |
| 15 |
Hb_013399_090 |
0.2053975019 |
- |
- |
PREDICTED: phospholipid:diacylglycerol acyltransferase 1-like [Jatropha curcas] |
| 16 |
Hb_007576_150 |
0.2076646609 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g57430, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000320_360 |
0.2085101531 |
- |
- |
PREDICTED: starch synthase 3, chloroplastic/amyloplastic isoform X1 [Jatropha curcas] |
| 18 |
Hb_080761_010 |
0.210378684 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 19 |
Hb_000340_360 |
0.2130389806 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase RKF3 [Jatropha curcas] |
| 20 |
Hb_001408_120 |
0.2153683003 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |